Remapping a dataset#
In this tutorial we will remap a dataset from the native ICON grid to HEALPix. The example dataset consists of five 2D variables sampled at six-hourly intervals.
Note
Please note that all of the methods used in this example are generic and can be applied to any combination of source and target grids.
import healpix as hp
import intake
import numpy as np
import xarray as xr
import easygems.healpix as egh
import easygems.remap as egr
cat = intake.open_catalog("https://data.nextgems-h2020.eu/online.yaml")
ds = cat.tutorial["ICON.native.2d_PT6H_inst"](chunks={}).to_dask()
ds
<xarray.Dataset> Size: 3GB
Dimensions: (time: 124, ncells: 1310720)
Coordinates:
* time (time) datetime64[ns] 992B 2020-01-01T05:59:59 ... 20...
cell_sea_land_mask (ncells) float64 10MB dask.array<chunksize=(1310720,), meta=np.ndarray>
lat (ncells) float64 10MB dask.array<chunksize=(1310720,), meta=np.ndarray>
lon (ncells) float64 10MB dask.array<chunksize=(1310720,), meta=np.ndarray>
Dimensions without coordinates: ncells
Data variables:
hus2m (time, ncells) float32 650MB dask.array<chunksize=(1, 1310720), meta=np.ndarray>
psl (time, ncells) float32 650MB dask.array<chunksize=(1, 1310720), meta=np.ndarray>
ts (time, ncells) float32 650MB dask.array<chunksize=(1, 1310720), meta=np.ndarray>
uas (time, ncells) float32 650MB dask.array<chunksize=(1, 1310720), meta=np.ndarray>
vas (time, ncells) float32 650MB dask.array<chunksize=(1, 1310720), meta=np.ndarray>- time: 124
- ncells: 1310720
- time(time)datetime64[ns]2020-01-01T05:59:59 ... 2020-01-...
array(['2020-01-01T05:59:59.000000000', '2020-01-01T11:59:59.000000000', '2020-01-01T17:59:59.000000000', '2020-01-01T23:59:59.000000000', '2020-01-02T05:59:59.000000000', '2020-01-02T11:59:59.000000000', '2020-01-02T17:59:59.000000000', '2020-01-02T23:59:59.000000000', '2020-01-03T05:59:59.000000000', '2020-01-03T11:59:59.000000000', '2020-01-03T17:59:59.000000000', '2020-01-03T23:59:59.000000000', '2020-01-04T05:59:59.000000000', '2020-01-04T11:59:59.000000000', '2020-01-04T17:59:59.000000000', '2020-01-04T23:59:59.000000000', '2020-01-05T05:59:59.000000000', '2020-01-05T11:59:59.000000000', '2020-01-05T17:59:59.000000000', '2020-01-05T23:59:59.000000000', '2020-01-06T05:59:59.000000000', '2020-01-06T11:59:59.000000000', '2020-01-06T17:59:59.000000000', '2020-01-06T23:59:59.000000000', '2020-01-07T05:59:59.000000000', '2020-01-07T11:59:59.000000000', '2020-01-07T17:59:59.000000000', '2020-01-07T23:59:59.000000000', '2020-01-08T05:59:59.000000000', '2020-01-08T11:59:59.000000000', '2020-01-08T17:59:59.000000000', '2020-01-08T23:59:59.000000000', '2020-01-09T05:59:59.000000000', '2020-01-09T11:59:59.000000000', '2020-01-09T17:59:59.000000000', '2020-01-09T23:59:59.000000000', '2020-01-10T05:59:59.000000000', '2020-01-10T11:59:59.000000000', '2020-01-10T17:59:59.000000000', '2020-01-10T23:59:59.000000000', '2020-01-11T05:59:59.000000000', '2020-01-11T11:59:59.000000000', '2020-01-11T17:59:59.000000000', '2020-01-11T23:59:59.000000000', '2020-01-12T05:59:59.000000000', '2020-01-12T11:59:59.000000000', '2020-01-12T17:59:59.000000000', '2020-01-12T23:59:59.000000000', '2020-01-13T05:59:59.000000000', '2020-01-13T11:59:59.000000000', '2020-01-13T17:59:59.000000000', '2020-01-13T23:59:59.000000000', '2020-01-14T05:59:59.000000000', '2020-01-14T11:59:59.000000000', '2020-01-14T17:59:59.000000000', '2020-01-14T23:59:59.000000000', '2020-01-15T05:59:59.000000000', '2020-01-15T11:59:59.000000000', '2020-01-15T17:59:59.000000000', '2020-01-15T23:59:59.000000000', '2020-01-16T05:59:59.000000000', '2020-01-16T11:59:59.000000000', '2020-01-16T17:59:59.000000000', '2020-01-16T23:59:59.000000000', '2020-01-17T05:59:59.000000000', '2020-01-17T11:59:59.000000000', '2020-01-17T17:59:59.000000000', '2020-01-17T23:59:59.000000000', '2020-01-18T05:59:59.000000000', '2020-01-18T11:59:59.000000000', '2020-01-18T17:59:59.000000000', '2020-01-18T23:59:59.000000000', '2020-01-19T05:59:59.000000000', '2020-01-19T11:59:59.000000000', '2020-01-19T17:59:59.000000000', '2020-01-19T23:59:59.000000000', '2020-01-20T05:59:59.000000000', '2020-01-20T11:59:59.000000000', '2020-01-20T17:59:59.000000000', '2020-01-20T23:59:59.000000000', '2020-01-21T05:59:59.000000000', '2020-01-21T11:59:59.000000000', '2020-01-21T17:59:59.000000000', '2020-01-21T23:59:59.000000000', '2020-01-22T05:59:59.000000000', '2020-01-22T11:59:59.000000000', '2020-01-22T17:59:59.000000000', '2020-01-22T23:59:59.000000000', '2020-01-23T05:59:59.000000000', '2020-01-23T11:59:59.000000000', '2020-01-23T17:59:59.000000000', '2020-01-23T23:59:59.000000000', '2020-01-24T05:59:59.000000000', '2020-01-24T11:59:59.000000000', '2020-01-24T17:59:59.000000000', '2020-01-24T23:59:59.000000000', '2020-01-25T05:59:59.000000000', '2020-01-25T11:59:59.000000000', '2020-01-25T17:59:59.000000000', '2020-01-25T23:59:59.000000000', '2020-01-26T05:59:59.000000000', '2020-01-26T11:59:59.000000000', '2020-01-26T17:59:59.000000000', '2020-01-26T23:59:59.000000000', '2020-01-27T05:59:59.000000000', '2020-01-27T11:59:59.000000000', '2020-01-27T17:59:59.000000000', '2020-01-27T23:59:59.000000000', '2020-01-28T05:59:59.000000000', '2020-01-28T11:59:59.000000000', '2020-01-28T17:59:59.000000000', '2020-01-28T23:59:59.000000000', '2020-01-29T05:59:59.000000000', '2020-01-29T11:59:59.000000000', '2020-01-29T17:59:59.000000000', '2020-01-29T23:59:59.000000000', '2020-01-30T05:59:59.000000000', '2020-01-30T11:59:59.000000000', '2020-01-30T17:59:59.000000000', '2020-01-30T23:59:59.000000000', '2020-01-31T05:59:59.000000000', '2020-01-31T11:59:59.000000000', '2020-01-31T17:59:59.000000000', '2020-01-31T23:59:59.000000000'], dtype='datetime64[ns]') - cell_sea_land_mask(ncells)float64dask.array<chunksize=(1310720,), meta=np.ndarray>
- long_name :
- sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell
- units :
- 2,1,-1,-
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
- dtype :
- int32
- zlib :
- False
- szip :
- False
- zstd :
- False
- bzip2 :
- False
- blosc :
- False
- shuffle :
- False
- complevel :
- 0
- fletcher32 :
- False
- contiguous :
- True
- chunksizes :
- None
- source :
- /work/bm1344/DKRZ/kerchunks_batched/erc2002/icon_grid_0033_R02B08_G.nc
- original_shape :
- [5242880]
Array Chunk Bytes 10.00 MiB 10.00 MiB Shape (1310720,) (1310720,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lat(ncells)float64dask.array<chunksize=(1310720,), meta=np.ndarray>
Array Chunk Bytes 10.00 MiB 10.00 MiB Shape (1310720,) (1310720,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lon(ncells)float64dask.array<chunksize=(1310720,), meta=np.ndarray>
Array Chunk Bytes 10.00 MiB 10.00 MiB Shape (1310720,) (1310720,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray
- hus2m(time, ncells)float32dask.array<chunksize=(1, 1310720), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- specific humidity in 2m
- number_of_grid_in_reference :
- 1
- param :
- 6.0.0
- standard_name :
- qv2m
- units :
- kg kg-1
Array Chunk Bytes 620.00 MiB 5.00 MiB Shape (124, 1310720) (1, 1310720) Dask graph 124 chunks in 2 graph layers Data type float32 numpy.ndarray - psl(time, ncells)float32dask.array<chunksize=(1, 1310720), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- mean sea level pressure
- number_of_grid_in_reference :
- 1
- param :
- 1.3.0
- standard_name :
- mean sea level pressure
- units :
- Pa
Array Chunk Bytes 620.00 MiB 5.00 MiB Shape (124, 1310720) (1, 1310720) Dask graph 124 chunks in 2 graph layers Data type float32 numpy.ndarray - ts(time, ncells)float32dask.array<chunksize=(1, 1310720), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- surface temperature
- number_of_grid_in_reference :
- 1
- param :
- 0.0.0
- standard_name :
- surface_temperature
- units :
- K
Array Chunk Bytes 620.00 MiB 5.00 MiB Shape (124, 1310720) (1, 1310720) Dask graph 124 chunks in 2 graph layers Data type float32 numpy.ndarray - uas(time, ncells)float32dask.array<chunksize=(1, 1310720), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- zonal wind in 10m
- number_of_grid_in_reference :
- 1
- param :
- 2.2.0
- standard_name :
- uas
- units :
- m s-1
Array Chunk Bytes 620.00 MiB 5.00 MiB Shape (124, 1310720) (1, 1310720) Dask graph 124 chunks in 2 graph layers Data type float32 numpy.ndarray - vas(time, ncells)float32dask.array<chunksize=(1, 1310720), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- meridional wind in 10m
- number_of_grid_in_reference :
- 1
- param :
- 3.2.0
- standard_name :
- vas
- units :
- m s-1
Array Chunk Bytes 620.00 MiB 5.00 MiB Shape (124, 1310720) (1, 1310720) Dask graph 124 chunks in 2 graph layers Data type float32 numpy.ndarray
The first step is to create a HEALPix grid that is close to the resolution of our source grid. Here we will choose an order of 8 (also known as zoom or refinement level) and nested ordering.
order = zoom = 8
nside = hp.order2nside(order)
pixels = np.arange(hp.nside2npix(nside))
hp_lon, hp_lat = hp.pix2ang(nside=nside, ipix=pixels, lonlat=True, nest=True)
Next, we can use our defined source and target grids to compute interpolation weights.
The easygems package provides a function to compute these weights using the Delaunay triangulation method.
weights = egr.compute_weights_delaunay(
points=(ds.lon, ds.lat),
xi=(hp_lon, hp_lat)
)
Tip
For larger grids, computing the remapping weights can be quite resource-intensive. However, you can save the calculated weights for future use:
weights.to_netcdf("healpix_weights.nc")
These weights can be applied to single fields directly:
egh.healpix_show(egr.apply_weights(ds.ts.isel(time=0), **weights));
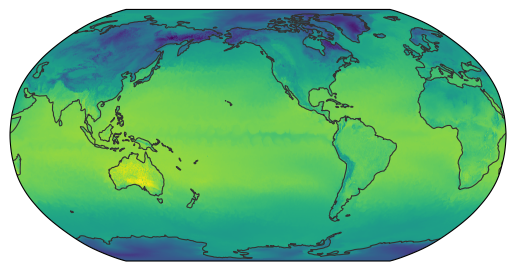
Alternatively, we can use xarray’s apply_ufunc() function to lift the function onto a full dataset. This requires a coupled of additional information from the user, e.g. the input dimension along which the function should be applied, and the resulting output dimensions name and size.
ds_remap = xr.apply_ufunc(
egr.apply_weights,
ds,
kwargs=weights,
keep_attrs=True,
input_core_dims=[["ncells"]],
output_core_dims=[["cell"]],
output_dtypes=["f4"],
vectorize=True,
dask="parallelized",
dask_gufunc_kwargs={
"output_sizes": {"cell": weights.sizes["tgt_idx"]},
},
)
ds_remap
<xarray.Dataset> Size: 2GB
Dimensions: (time: 124, cell: 786432)
Coordinates:
* time (time) datetime64[ns] 992B 2020-01-01T05:59:59 ... 2020-01-31T23...
Dimensions without coordinates: cell
Data variables:
hus2m (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
psl (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
ts (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
uas (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
vas (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>- time: 124
- cell: 786432
- time(time)datetime64[ns]2020-01-01T05:59:59 ... 2020-01-...
array(['2020-01-01T05:59:59.000000000', '2020-01-01T11:59:59.000000000', '2020-01-01T17:59:59.000000000', '2020-01-01T23:59:59.000000000', '2020-01-02T05:59:59.000000000', '2020-01-02T11:59:59.000000000', '2020-01-02T17:59:59.000000000', '2020-01-02T23:59:59.000000000', '2020-01-03T05:59:59.000000000', '2020-01-03T11:59:59.000000000', '2020-01-03T17:59:59.000000000', '2020-01-03T23:59:59.000000000', '2020-01-04T05:59:59.000000000', '2020-01-04T11:59:59.000000000', '2020-01-04T17:59:59.000000000', '2020-01-04T23:59:59.000000000', '2020-01-05T05:59:59.000000000', '2020-01-05T11:59:59.000000000', '2020-01-05T17:59:59.000000000', '2020-01-05T23:59:59.000000000', '2020-01-06T05:59:59.000000000', '2020-01-06T11:59:59.000000000', '2020-01-06T17:59:59.000000000', '2020-01-06T23:59:59.000000000', '2020-01-07T05:59:59.000000000', '2020-01-07T11:59:59.000000000', '2020-01-07T17:59:59.000000000', '2020-01-07T23:59:59.000000000', '2020-01-08T05:59:59.000000000', '2020-01-08T11:59:59.000000000', '2020-01-08T17:59:59.000000000', '2020-01-08T23:59:59.000000000', '2020-01-09T05:59:59.000000000', '2020-01-09T11:59:59.000000000', '2020-01-09T17:59:59.000000000', '2020-01-09T23:59:59.000000000', '2020-01-10T05:59:59.000000000', '2020-01-10T11:59:59.000000000', '2020-01-10T17:59:59.000000000', '2020-01-10T23:59:59.000000000', '2020-01-11T05:59:59.000000000', '2020-01-11T11:59:59.000000000', '2020-01-11T17:59:59.000000000', '2020-01-11T23:59:59.000000000', '2020-01-12T05:59:59.000000000', '2020-01-12T11:59:59.000000000', '2020-01-12T17:59:59.000000000', '2020-01-12T23:59:59.000000000', '2020-01-13T05:59:59.000000000', '2020-01-13T11:59:59.000000000', '2020-01-13T17:59:59.000000000', '2020-01-13T23:59:59.000000000', '2020-01-14T05:59:59.000000000', '2020-01-14T11:59:59.000000000', '2020-01-14T17:59:59.000000000', '2020-01-14T23:59:59.000000000', '2020-01-15T05:59:59.000000000', '2020-01-15T11:59:59.000000000', '2020-01-15T17:59:59.000000000', '2020-01-15T23:59:59.000000000', '2020-01-16T05:59:59.000000000', '2020-01-16T11:59:59.000000000', '2020-01-16T17:59:59.000000000', '2020-01-16T23:59:59.000000000', '2020-01-17T05:59:59.000000000', '2020-01-17T11:59:59.000000000', '2020-01-17T17:59:59.000000000', '2020-01-17T23:59:59.000000000', '2020-01-18T05:59:59.000000000', '2020-01-18T11:59:59.000000000', '2020-01-18T17:59:59.000000000', '2020-01-18T23:59:59.000000000', '2020-01-19T05:59:59.000000000', '2020-01-19T11:59:59.000000000', '2020-01-19T17:59:59.000000000', '2020-01-19T23:59:59.000000000', '2020-01-20T05:59:59.000000000', '2020-01-20T11:59:59.000000000', '2020-01-20T17:59:59.000000000', '2020-01-20T23:59:59.000000000', '2020-01-21T05:59:59.000000000', '2020-01-21T11:59:59.000000000', '2020-01-21T17:59:59.000000000', '2020-01-21T23:59:59.000000000', '2020-01-22T05:59:59.000000000', '2020-01-22T11:59:59.000000000', '2020-01-22T17:59:59.000000000', '2020-01-22T23:59:59.000000000', '2020-01-23T05:59:59.000000000', '2020-01-23T11:59:59.000000000', '2020-01-23T17:59:59.000000000', '2020-01-23T23:59:59.000000000', '2020-01-24T05:59:59.000000000', '2020-01-24T11:59:59.000000000', '2020-01-24T17:59:59.000000000', '2020-01-24T23:59:59.000000000', '2020-01-25T05:59:59.000000000', '2020-01-25T11:59:59.000000000', '2020-01-25T17:59:59.000000000', '2020-01-25T23:59:59.000000000', '2020-01-26T05:59:59.000000000', '2020-01-26T11:59:59.000000000', '2020-01-26T17:59:59.000000000', '2020-01-26T23:59:59.000000000', '2020-01-27T05:59:59.000000000', '2020-01-27T11:59:59.000000000', '2020-01-27T17:59:59.000000000', '2020-01-27T23:59:59.000000000', '2020-01-28T05:59:59.000000000', '2020-01-28T11:59:59.000000000', '2020-01-28T17:59:59.000000000', '2020-01-28T23:59:59.000000000', '2020-01-29T05:59:59.000000000', '2020-01-29T11:59:59.000000000', '2020-01-29T17:59:59.000000000', '2020-01-29T23:59:59.000000000', '2020-01-30T05:59:59.000000000', '2020-01-30T11:59:59.000000000', '2020-01-30T17:59:59.000000000', '2020-01-30T23:59:59.000000000', '2020-01-31T05:59:59.000000000', '2020-01-31T11:59:59.000000000', '2020-01-31T17:59:59.000000000', '2020-01-31T23:59:59.000000000'], dtype='datetime64[ns]')
- hus2m(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- specific humidity in 2m
- number_of_grid_in_reference :
- 1
- param :
- 6.0.0
- standard_name :
- qv2m
- units :
- kg kg-1
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - psl(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- mean sea level pressure
- number_of_grid_in_reference :
- 1
- param :
- 1.3.0
- standard_name :
- mean sea level pressure
- units :
- Pa
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - ts(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- surface temperature
- number_of_grid_in_reference :
- 1
- param :
- 0.0.0
- standard_name :
- surface_temperature
- units :
- K
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - uas(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- zonal wind in 10m
- number_of_grid_in_reference :
- 1
- param :
- 2.2.0
- standard_name :
- uas
- units :
- m s-1
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - vas(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- meridional wind in 10m
- number_of_grid_in_reference :
- 1
- param :
- 3.2.0
- standard_name :
- vas
- units :
- m s-1
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray
The resulting dataset can then be used as usual and the remapping is performed on demand.
egh.healpix_show(ds_remap.ts.isel(time=0));
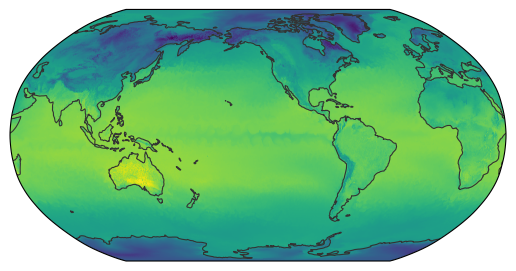
Attaching a coordinate reference system#
It is good practice to store map projection information in the Coordinate Reference Systems (CRS).
By making the crs coordinate non-dimensional it will “stick” to the dataset even if individual variables are subselected.
crs = xr.DataArray(
name="crs",
attrs={
"grid_mapping_name": "healpix",
"refinement_level": order,
"indexing_scheme": "nested",
},
)
cell = xr.DataArray(
pixels, dims=("cell",), attrs={"standard_name": "healpix_index"}
)
ds_remap_cf = ds_remap.assign_coords(crs=crs, cell=cell)
ds_remap_cf
<xarray.Dataset> Size: 2GB
Dimensions: (time: 124, cell: 786432)
Coordinates:
* time (time) datetime64[ns] 992B 2020-01-01T05:59:59 ... 2020-01-31T23...
* cell (cell) int64 6MB 0 1 2 3 4 5 ... 786427 786428 786429 786430 786431
crs float64 8B nan
Data variables:
hus2m (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
psl (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
ts (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
uas (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>
vas (time, cell) float32 390MB dask.array<chunksize=(1, 786432), meta=np.ndarray>- time: 124
- cell: 786432
- time(time)datetime64[ns]2020-01-01T05:59:59 ... 2020-01-...
array(['2020-01-01T05:59:59.000000000', '2020-01-01T11:59:59.000000000', '2020-01-01T17:59:59.000000000', '2020-01-01T23:59:59.000000000', '2020-01-02T05:59:59.000000000', '2020-01-02T11:59:59.000000000', '2020-01-02T17:59:59.000000000', '2020-01-02T23:59:59.000000000', '2020-01-03T05:59:59.000000000', '2020-01-03T11:59:59.000000000', '2020-01-03T17:59:59.000000000', '2020-01-03T23:59:59.000000000', '2020-01-04T05:59:59.000000000', '2020-01-04T11:59:59.000000000', '2020-01-04T17:59:59.000000000', '2020-01-04T23:59:59.000000000', '2020-01-05T05:59:59.000000000', '2020-01-05T11:59:59.000000000', '2020-01-05T17:59:59.000000000', '2020-01-05T23:59:59.000000000', '2020-01-06T05:59:59.000000000', '2020-01-06T11:59:59.000000000', '2020-01-06T17:59:59.000000000', '2020-01-06T23:59:59.000000000', '2020-01-07T05:59:59.000000000', '2020-01-07T11:59:59.000000000', '2020-01-07T17:59:59.000000000', '2020-01-07T23:59:59.000000000', '2020-01-08T05:59:59.000000000', '2020-01-08T11:59:59.000000000', '2020-01-08T17:59:59.000000000', '2020-01-08T23:59:59.000000000', '2020-01-09T05:59:59.000000000', '2020-01-09T11:59:59.000000000', '2020-01-09T17:59:59.000000000', '2020-01-09T23:59:59.000000000', '2020-01-10T05:59:59.000000000', '2020-01-10T11:59:59.000000000', '2020-01-10T17:59:59.000000000', '2020-01-10T23:59:59.000000000', '2020-01-11T05:59:59.000000000', '2020-01-11T11:59:59.000000000', '2020-01-11T17:59:59.000000000', '2020-01-11T23:59:59.000000000', '2020-01-12T05:59:59.000000000', '2020-01-12T11:59:59.000000000', '2020-01-12T17:59:59.000000000', '2020-01-12T23:59:59.000000000', '2020-01-13T05:59:59.000000000', '2020-01-13T11:59:59.000000000', '2020-01-13T17:59:59.000000000', '2020-01-13T23:59:59.000000000', '2020-01-14T05:59:59.000000000', '2020-01-14T11:59:59.000000000', '2020-01-14T17:59:59.000000000', '2020-01-14T23:59:59.000000000', '2020-01-15T05:59:59.000000000', '2020-01-15T11:59:59.000000000', '2020-01-15T17:59:59.000000000', '2020-01-15T23:59:59.000000000', '2020-01-16T05:59:59.000000000', '2020-01-16T11:59:59.000000000', '2020-01-16T17:59:59.000000000', '2020-01-16T23:59:59.000000000', '2020-01-17T05:59:59.000000000', '2020-01-17T11:59:59.000000000', '2020-01-17T17:59:59.000000000', '2020-01-17T23:59:59.000000000', '2020-01-18T05:59:59.000000000', '2020-01-18T11:59:59.000000000', '2020-01-18T17:59:59.000000000', '2020-01-18T23:59:59.000000000', '2020-01-19T05:59:59.000000000', '2020-01-19T11:59:59.000000000', '2020-01-19T17:59:59.000000000', '2020-01-19T23:59:59.000000000', '2020-01-20T05:59:59.000000000', '2020-01-20T11:59:59.000000000', '2020-01-20T17:59:59.000000000', '2020-01-20T23:59:59.000000000', '2020-01-21T05:59:59.000000000', '2020-01-21T11:59:59.000000000', '2020-01-21T17:59:59.000000000', '2020-01-21T23:59:59.000000000', '2020-01-22T05:59:59.000000000', '2020-01-22T11:59:59.000000000', '2020-01-22T17:59:59.000000000', '2020-01-22T23:59:59.000000000', '2020-01-23T05:59:59.000000000', '2020-01-23T11:59:59.000000000', '2020-01-23T17:59:59.000000000', '2020-01-23T23:59:59.000000000', '2020-01-24T05:59:59.000000000', '2020-01-24T11:59:59.000000000', '2020-01-24T17:59:59.000000000', '2020-01-24T23:59:59.000000000', '2020-01-25T05:59:59.000000000', '2020-01-25T11:59:59.000000000', '2020-01-25T17:59:59.000000000', '2020-01-25T23:59:59.000000000', '2020-01-26T05:59:59.000000000', '2020-01-26T11:59:59.000000000', '2020-01-26T17:59:59.000000000', '2020-01-26T23:59:59.000000000', '2020-01-27T05:59:59.000000000', '2020-01-27T11:59:59.000000000', '2020-01-27T17:59:59.000000000', '2020-01-27T23:59:59.000000000', '2020-01-28T05:59:59.000000000', '2020-01-28T11:59:59.000000000', '2020-01-28T17:59:59.000000000', '2020-01-28T23:59:59.000000000', '2020-01-29T05:59:59.000000000', '2020-01-29T11:59:59.000000000', '2020-01-29T17:59:59.000000000', '2020-01-29T23:59:59.000000000', '2020-01-30T05:59:59.000000000', '2020-01-30T11:59:59.000000000', '2020-01-30T17:59:59.000000000', '2020-01-30T23:59:59.000000000', '2020-01-31T05:59:59.000000000', '2020-01-31T11:59:59.000000000', '2020-01-31T17:59:59.000000000', '2020-01-31T23:59:59.000000000'], dtype='datetime64[ns]') - cell(cell)int640 1 2 3 ... 786429 786430 786431
- standard_name :
- healpix_index
array([ 0, 1, 2, ..., 786429, 786430, 786431], shape=(786432,))
- crs()float64nan
- grid_mapping_name :
- healpix
- refinement_level :
- 8
- indexing_scheme :
- nested
array(nan)
- hus2m(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- specific humidity in 2m
- number_of_grid_in_reference :
- 1
- param :
- 6.0.0
- standard_name :
- qv2m
- units :
- kg kg-1
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - psl(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- mean sea level pressure
- number_of_grid_in_reference :
- 1
- param :
- 1.3.0
- standard_name :
- mean sea level pressure
- units :
- Pa
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - ts(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- surface temperature
- number_of_grid_in_reference :
- 1
- param :
- 0.0.0
- standard_name :
- surface_temperature
- units :
- K
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - uas(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- zonal wind in 10m
- number_of_grid_in_reference :
- 1
- param :
- 2.2.0
- standard_name :
- uas
- units :
- m s-1
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray - vas(time, cell)float32dask.array<chunksize=(1, 786432), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- long_name :
- meridional wind in 10m
- number_of_grid_in_reference :
- 1
- param :
- 3.2.0
- standard_name :
- vas
- units :
- m s-1
Array Chunk Bytes 372.00 MiB 3.00 MiB Shape (124, 786432) (1, 786432) Dask graph 124 chunks in 5 graph layers Data type float32 numpy.ndarray
