Grid Index Ordering#
Cell indices in ICON global grids are ordered in a specific manner which simplifies a bunch of operations an which is interesting to know about. Here, we’ll look at a few aspects of this particular ordering.
%matplotlib inline
import matplotlib.pylab as plt
import numpy as np
import xarray as xr
import cartopy.crs as ccrs
import cartopy.feature as cfeature
We’ll start using a relatively coarse (R02B03) grid in order to make the plots less crowded and our live easier.
grid = xr.open_dataset(
"/pool/data/ICON/grids/public/mpim/0030/icon_grid_0030_R02B03_G.nc"
)
grid
<xarray.Dataset>
Dimensions: (cell: 5120, nv: 3, vertex: 2562, ne: 6, edge: 7680, no: 4, nc: 2, max_stored_decompositions: 4, two_grf: 2, cell_grf: 14, max_chdom: 1, edge_grf: 24, vert_grf: 13)
Coordinates:
clon (cell) float64 1.274 1.34 ... 1.437 1.329
clat (cell) float64 0.9184 0.9398 ... -0.8052
vlon (vertex) float64 1.274 1.213 ... 1.274 1.325
vlat (vertex) float64 0.9625 0.8955 ... -0.7609
elon (edge) float64 1.306 1.242 ... 1.407 1.354
elat (edge) float64 0.9292 0.9292 ... -0.7939
Dimensions without coordinates: cell, nv, vertex, ne, edge, no, nc, max_stored_decompositions, two_grf, cell_grf, max_chdom, edge_grf, vert_grf
Data variables: (12/91)
clon_vertices (cell, nv) float64 1.274 1.213 ... 1.325
clat_vertices (cell, nv) float64 0.9625 0.8955 ... -0.7609
vlon_vertices (vertex, ne) float64 9.969e+36 ... 9.969e+36
vlat_vertices (vertex, ne) float64 9.969e+36 ... 9.969e+36
elon_vertices (edge, no) float64 1.335 1.34 ... 1.385
elat_vertices (edge, no) float64 0.8955 0.9398 ... -0.8265
... ...
edge_dual_normal_cartesian_x (edge) float64 -0.2769 -0.6793 ... 0.6559
edge_dual_normal_cartesian_y (edge) float64 0.8043 -0.512 ... 0.4705
edge_dual_normal_cartesian_z (edge) float64 -0.5257 0.5257 ... 0.5902
cell_circumcenter_cartesian_x (cell) float64 0.1775 0.1351 ... 0.1662
cell_circumcenter_cartesian_y (cell) float64 0.5805 0.5743 ... 0.6727
cell_circumcenter_cartesian_z (cell) float64 0.7947 0.8074 ... -0.721
Attributes: (12/43)
title: ICON grid description
institution: Max Planck Institute for Meteorology/Deutscher ...
source: git@git.mpimet.mpg.de:GridGenerator.git
revision: 2cdf99bec403902989148ebb4bd38c218a64a1b3
date: 20190405 at 141502
user_name: Rene Redler (m300083)
... ...
topography: modified SRTM30
symmetry: along equator
subcentre: 1
number_of_grid_used: 30
ICON_grid_file_uri: http://icon-downloads.mpimet.mpg.de/grids/publi...
NCO: netCDF Operators version 4.7.5 (Homepage = http...- cell: 5120
- nv: 3
- vertex: 2562
- ne: 6
- edge: 7680
- no: 4
- nc: 2
- max_stored_decompositions: 4
- two_grf: 2
- cell_grf: 14
- max_chdom: 1
- edge_grf: 24
- vert_grf: 13
- clon(cell)float64...
- long_name :
- center longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- clon_vertices
array([1.27409 , 1.339784, 1.208396, ..., 1.37425 , 1.436983, 1.328586])
- clat(cell)float64...
- long_name :
- center latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- clat_vertices
array([ 0.918432, 0.93978 , 0.93978 , ..., -0.734887, -0.79992 , -0.805211])
- vlon(vertex)float64...
- long_name :
- vertex longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- vlon_vertices
array([1.27409 , 1.213028, 1.335153, ..., 1.222891, 1.27409 , 1.32529 ])
- vlat(vertex)float64...
- long_name :
- vertex latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- vlat_vertices
array([ 0.9625 , 0.895474, 0.895474, ..., -0.760894, -0.692827, -0.760894])
- elon(edge)float64...
- long_name :
- edge midpoint longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- elon_vertices
array([1.305992, 1.242189, 1.27409 , ..., 1.376351, 1.407033, 1.354338])
- elat(edge)float64...
- long_name :
- edge midpoint latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- elat_vertices
array([ 0.92921 , 0.92921 , 0.896384, ..., -0.759171, -0.79145 , -0.793938])
- clon_vertices(cell, nv)float64...
- units :
- radian
array([[1.27409 , 1.213028, 1.335153], [1.335153, 1.408679, 1.27409 ], [1.27409 , 1.139502, 1.213028], ..., [1.32529 , 1.368626, 1.427183], [1.427183, 1.49535 , 1.385393], [1.385393, 1.27409 , 1.32529 ]]) - clat_vertices(cell, nv)float64...
- units :
- radian
array([[ 0.9625 , 0.895474, 0.895474], [ 0.895474, 0.958823, 0.9625 ], [ 0.9625 , 0.958823, 0.895474], ..., [-0.760894, -0.69077 , -0.756151], [-0.756151, -0.818425, -0.826531], [-0.826531, -0.829283, -0.760894]]) - vlon_vertices(vertex, ne)float64...
- units :
- radian
array([[9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], ..., [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36]]) - vlat_vertices(vertex, ne)float64...
- units :
- radian
array([[9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], ..., [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36], [9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36, 9.96921e+36]]) - elon_vertices(edge, no)float64...
- units :
- radian
array([[1.335153, 1.339784, 1.27409 , 1.27409 ], [1.27409 , 1.208396, 1.213028, 1.27409 ], [1.213028, 1.27409 , 1.335153, 1.27409 ], ..., [1.378493, 1.427183, 1.37425 , 1.32529 ], [1.378493, 1.385393, 1.436983, 1.427183], [1.378493, 1.32529 , 1.328586, 1.385393]]) - elat_vertices(edge, no)float64...
- units :
- radian
array([[ 0.895474, 0.93978 , 0.9625 , 0.918432], [ 0.9625 , 0.93978 , 0.895474, 0.918432], [ 0.895474, 0.873682, 0.895474, 0.918432], ..., [-0.782817, -0.756151, -0.734887, -0.760894], [-0.782817, -0.826531, -0.79992 , -0.756151], [-0.782817, -0.760894, -0.805211, -0.826531]]) - ifs2icon_cell_grid(cell)float64...
- long_name :
- ifs to icon cells
array([9.96921e+36, 9.96921e+36, 9.96921e+36, ..., 9.96921e+36, 9.96921e+36, 9.96921e+36]) - ifs2icon_edge_grid(edge)float64...
- long_name :
- ifs to icon edge
array([9.96921e+36, 9.96921e+36, 9.96921e+36, ..., 9.96921e+36, 9.96921e+36, 9.96921e+36]) - ifs2icon_vertex_grid(vertex)float64...
- long_name :
- ifs to icon vertex
array([9.96921e+36, 9.96921e+36, 9.96921e+36, ..., 9.96921e+36, 9.96921e+36, 9.96921e+36]) - cell_area(cell)float64...
- long_name :
- area of grid cell
- units :
- m2
- standard_name :
- area
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([1.024614e+11, 1.039760e+11, 1.039760e+11, ..., 1.032592e+11, 1.034199e+11, 1.037767e+11]) - dual_area(vertex)float64...
- long_name :
- areas of dual hexagonal/pentagonal cells
- units :
- m2
- standard_name :
- area
array([2.061924e+11, 2.062077e+11, 2.062077e+11, ..., 2.051674e+11, 2.042820e+11, 2.051674e+11]) - phys_cell_id(cell)int32...
- long_name :
- physical domain ID of cell
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([1, 1, 1, ..., 1, 1, 1], dtype=int32)
- phys_edge_id(edge)int32...
- long_name :
- physical domain ID of edge
array([1, 1, 1, ..., 1, 1, 1], dtype=int32)
- lon_cell_centre(cell)float64...
- long_name :
- longitude of cell centre
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([1.27409 , 1.339784, 1.208396, ..., 1.37425 , 1.436983, 1.328586])
- lat_cell_centre(cell)float64...
- long_name :
- latitude of cell centre
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([ 0.918432, 0.93978 , 0.93978 , ..., -0.734887, -0.79992 , -0.805211])
- lat_cell_barycenter(cell)float64...
- long_name :
- latitude of cell barycenter
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([ 0.91843 , 0.939636, 0.939636, ..., -0.736368, -0.800875, -0.806078])
- lon_cell_barycenter(cell)float64...
- long_name :
- longitude of cell barycenter
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([1.27409 , 1.339296, 1.208884, ..., 1.37367 , 1.435929, 1.328242])
- longitude_vertices(vertex)float64...
- long_name :
- longitude of vertices
- units :
- radian
array([1.27409 , 1.213028, 1.335153, ..., 1.222891, 1.27409 , 1.32529 ])
- latitude_vertices(vertex)float64...
- long_name :
- latitude of vertices
- units :
- radian
array([ 0.9625 , 0.895474, 0.895474, ..., -0.760894, -0.692827, -0.760894])
- lon_edge_centre(edge)float64...
- long_name :
- longitudes of edge midpoints
- units :
- radian
array([1.305992, 1.242189, 1.27409 , ..., 1.376351, 1.407033, 1.354338])
- lat_edge_centre(edge)float64...
- long_name :
- latitudes of edge midpoints
- units :
- radian
array([ 0.92921 , 0.92921 , 0.896384, ..., -0.759171, -0.79145 , -0.793938])
- edge_of_cell(nv, cell)int32...
- long_name :
- edges of each cellvertices
array([[ 1, 5, 6, ..., 7669, 7363, 7623], [ 2, 4, 7, ..., 7364, 7624, 7671], [ 3, 1, 2, ..., 7678, 7679, 7680]], dtype=int32) - vertex_of_cell(nv, cell)int32...
- long_name :
- vertices of each cellcells ad
array([[ 1, 3, 1, ..., 2562, 2475, 2552], [ 2, 4, 5, ..., 2474, 2470, 2547], [ 3, 1, 2, ..., 2475, 2552, 2562]], dtype=int32) - adjacent_cell_of_edge(nc, edge)int32...
- long_name :
- cells adjacent to each edge
array([[ 1, 1, 1, ..., 5117, 5117, 5117], [ 2, 3, 4, ..., 5118, 5119, 5120]], dtype=int32) - edge_vertices(nc, edge)int32...
- long_name :
- vertices at the end of of each edge
array([[ 3, 1, 2, ..., 2562, 2475, 2552], [ 1, 2, 3, ..., 2475, 2552, 2562]], dtype=int32) - cells_of_vertex(ne, vertex)int32...
- long_name :
- cells around each vertex
array([[ 1, 1, 1, ..., 5105, 5105, 5105], [ 2, 3, 2, ..., 5106, 5106, 5107], [ 3, 4, 4, ..., 5108, 5107, 5108], [ 5, 9, 13, ..., 5109, 5113, 5117], [ 6, 10, 14, ..., 5110, 5114, 5118], [ 8, 12, 16, ..., 5112, 5116, 5120]], dtype=int32) - edges_of_vertex(ne, vertex)int32...
- long_name :
- edges around each vertex
array([[ 1, 2, 1, ..., 7663, 7663, 7664], [ 2, 3, 3, ..., 7665, 7664, 7665], [ 4, 7, 5, ..., 7666, 7667, 7669], [ 6, 8, 9, ..., 7670, 7668, 7671], [ 10, 17, 24, ..., 7672, 7675, 7678], [ 12, 19, 26, ..., 7674, 7677, 7680]], dtype=int32) - vertices_of_vertex(ne, vertex)int32...
- long_name :
- vertices around each vertex
array([[ 3, 1, 1, ..., 2561, 2560, 2561], [ 2, 3, 2, ..., 2562, 2562, 2560], [ 4, 5, 4, ..., 2511, 2511, 2474], [ 5, 6, 6, ..., 2547, 2474, 2547], [ 7, 10, 13, ..., 2551, 2512, 2475], [ 8, 11, 14, ..., 2513, 2476, 2552]], dtype=int32) - cell_area_p(cell)float64...
- long_name :
- area of grid cell
- units :
- m2
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([1.024614e+11, 1.039760e+11, 1.039760e+11, ..., 1.032592e+11, 1.034199e+11, 1.037767e+11]) - cell_elevation(cell)float64...
- long_name :
- elevation at the cell centers
- units :
- m
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([0., 0., 0., ..., 0., 0., 0.])
- cell_sea_land_mask(cell)int32...
- long_name :
- sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell
- units :
- 2,1,-1,-
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
array([0, 0, 0, ..., 0, 0, 0], dtype=int32)
- cell_domain_id(cell, max_stored_decompositions)int32...
- long_name :
- cell domain id for decomposition
array([[-1, -1, -1, -1], [-1, -1, -1, -1], [-1, -1, -1, -1], ..., [-1, -1, -1, -1], [-1, -1, -1, -1], [-1, -1, -1, -1]], dtype=int32) - cell_no_of_domains(max_stored_decompositions)int32...
- long_name :
- number of domains for each decomposition
array([0, 0, 0, 0], dtype=int32)
- dual_area_p(vertex)float64...
- long_name :
- areas of dual hexagonal/pentagonal cells
- units :
- m2
array([2.061924e+11, 2.062077e+11, 2.062077e+11, ..., 2.051674e+11, 2.042820e+11, 2.051674e+11]) - edge_length(edge)float64...
- long_name :
- lengths of edges of triangular cells
- units :
- m
array([486276.989186, 486276.989186, 486236.548474, ..., 472083.45348 , 485826.624142, 496849.001131]) - edge_cell_distance(nc, edge)float64...
- long_name :
- distances between edge midpoint and adjacent triangle midpoints
- units :
- m
array([[140440.067587, 140440.067587, 140475.07982 , ..., 150968.997 , 139640.532359, 129578.870499], [144572.866883, 144572.866883, 144639.495821, ..., 155033.252937, 144023.003658, 135041.419206]]) - dual_edge_length(edge)float64...
- long_name :
- lengths of dual edges (distances between triangular cell circumcenters)
- units :
- m
array([285012.93447 , 285012.93447 , 285114.575641, ..., 306002.249937, 283663.536017, 264620.289705]) - edgequad_area(edge)float64...
- long_name :
- area around the edge formed by the two adjacent triangles
- units :
- m2
array([0.001707, 0.001707, 0.001708, ..., 0.001779, 0.001697, 0.001619])
- edge_elevation(edge)float64...
- long_name :
- elevation at the edge centers
- units :
- m
array([0., 0., 0., ..., 0., 0., 0.])
- edge_sea_land_mask(edge)int32...
- long_name :
- sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell
- units :
- 2,1,-1,-
array([0, 0, 0, ..., 0, 0, 0], dtype=int32)
- edge_vert_distance(nc, edge)float64...
- long_name :
- distances between edge midpoint and vertices of that edge
- units :
- m
array([[243138.494593, 243138.494593, 243118.274237, ..., 236041.72674 , 242913.312071, 248424.500565], [243138.494593, 243138.494593, 243118.274237, ..., 236041.72674 , 242913.312071, 248424.500565]]) - zonal_normal_primal_edge(edge)float64...
- long_name :
- zonal component of normal to primal edge
- units :
- radian
array([ 8.784942e-01, -8.784942e-01, -7.187471e-16, ..., -6.406706e-02, 9.231028e-01, -8.419251e-01]) - meridional_normal_primal_edge(edge)float64...
- long_name :
- meridional component of normal to primal edge
- units :
- radian
array([ 0.477753, 0.477753, -1. , ..., 0.997946, -0.384553, -0.539594])
- zonal_normal_dual_edge(edge)float64...
- long_name :
- zonal component of normal to dual edge
- units :
- radian
array([ 0.477753, 0.477753, -1. , ..., 0.997946, -0.384553, -0.539594])
- meridional_normal_dual_edge(edge)float64...
- long_name :
- meridional component of normal to dual edge
- units :
- radian
array([-8.784942e-01, 8.784942e-01, 7.187471e-16, ..., 6.406706e-02, -9.231028e-01, 8.419251e-01]) - orientation_of_normal(nv, cell)int32...
- long_name :
- orientations of normals to triangular cell edges
array([[ 1, 1, 1, ..., -1, -1, -1], [ 1, 1, 1, ..., -1, -1, -1], [ 1, -1, -1, ..., -1, -1, -1]], dtype=int32) - cell_index(cell)int32...
- long_name :
- cell index
array([ 1, 2, 3, ..., 5118, 5119, 5120], dtype=int32)
- parent_cell_index(cell)int32...
- long_name :
- parent cell index
array([ 1, 1, 1, ..., 1280, 1280, 1280], dtype=int32)
- parent_cell_type(cell)int32...
- long_name :
- parent cell type
array([200, 203, 201, ..., 203, 201, 202], dtype=int32)
- neighbor_cell_index(nv, cell)int32...
- long_name :
- cell neighbor index
array([[ 2, 16, 8, ..., 5107, 4887, 5070], [ 3, 6, 10, ..., 4888, 5071, 5108], [ 4, 1, 1, ..., 5117, 5117, 5117]], dtype=int32) - child_cell_index(no, cell)int32...
- long_name :
- child cell index
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]], dtype=int32) - child_cell_id(cell)int32...
- long_name :
- domain ID of child cell
array([0, 0, 0, ..., 0, 0, 0], dtype=int32)
- edge_index(edge)int32...
- long_name :
- edge index
array([ 1, 2, 3, ..., 7678, 7679, 7680], dtype=int32)
- edge_parent_type(edge)int32...
- long_name :
- edge paren
array([202, 203, 201, ..., 202, 203, 201], dtype=int32)
- vertex_index(vertex)int32...
- long_name :
- vertices index
array([ 1, 2, 3, ..., 2560, 2561, 2562], dtype=int32)
- edge_orientation(ne, vertex)int32...
- long_name :
- edge orientation
array([[ 1, 1, -1, ..., 1, -1, -1], [-1, -1, 1, ..., -1, 1, 1], [ 1, 1, -1, ..., 1, -1, -1], [-1, -1, 1, ..., -1, 1, 1], [-1, -1, -1, ..., 1, 1, 1], [ 1, 1, 1, ..., -1, -1, -1]], dtype=int32) - edge_system_orientation(edge)int32...
- long_name :
- edge system orientation
array([-1, -1, -1, ..., 1, 1, 1], dtype=int32)
- refin_c_ctrl(cell)int32...
- long_name :
- refinement control flag for cells
array([-4, -4, -4, ..., -4, -4, -4], dtype=int32)
- index_c_list(two_grf, cell_grf)int32...
- long_name :
- list of start and end indices for each refinement control level for cells
array([[-2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647], [-2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647]], dtype=int32) - start_idx_c(max_chdom, cell_grf)int32...
- long_name :
- list of start indices for each refinement control level for cells
array([[5121, 5121, 5121, 5121, 5121, 5121, 5121, 5121, 5121, 1, 1, 1, 1, 1]], dtype=int32) - end_idx_c(max_chdom, cell_grf)int32...
- long_name :
- list of end indices for each refinement control level for cells
array([[5120, 5120, 5120, 5120, 5120, 5120, 5120, 5120, 5120, 0, 0, 0, 0, 0]], dtype=int32) - refin_e_ctrl(edge)int32...
- long_name :
- refinement control flag for edges
array([-8, -8, -8, ..., -8, -8, -8], dtype=int32)
- index_e_list(two_grf, edge_grf)int32...
- long_name :
- list of start and end indices for each refinement control level for edges
array([[-2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647], [-2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647]], dtype=int32) - start_idx_e(max_chdom, edge_grf)int32...
- long_name :
- list of start indices for each refinement control level for edges
array([[7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 7681, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]], dtype=int32) - end_idx_e(max_chdom, edge_grf)int32...
- long_name :
- list of end indices for each refinement control level for edges
array([[7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 7680, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], dtype=int32) - refin_v_ctrl(vertex)int32...
- long_name :
- refinement control flag for vertices
array([0, 0, 0, ..., 0, 0, 0], dtype=int32)
- index_v_list(two_grf, vert_grf)int32...
- long_name :
- list of start and end indices for each refinement control level for vertices
array([[-2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647], [-2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647, -2147483647]], dtype=int32) - start_idx_v(max_chdom, vert_grf)int32...
- long_name :
- list of start indices for each refinement control level for vertices
array([[2563, 2563, 2563, 2563, 2563, 2563, 2563, 2563, 1, 1, 1, 1, 1]], dtype=int32) - end_idx_v(max_chdom, vert_grf)int32...
- long_name :
- list of end indices for each refinement control level for vertices
array([[2562, 2562, 2562, 2562, 2562, 2562, 2562, 2562, 0, 0, 0, 0, 0]], dtype=int32) - parent_edge_index(edge)int32...
- long_name :
- parent edge index
array([ 2, 3, 1, ..., 1911, 1920, 1849], dtype=int32)
- child_edge_index(no, edge)int32...
- long_name :
- child edge index
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]], dtype=int32) - child_edge_id(edge)int32...
- long_name :
- domain ID of child edge
array([0, 0, 0, ..., 0, 0, 0], dtype=int32)
- parent_vertex_index(vertex)int32...
- long_name :
- parent vertex index
array([ -1, -2, -3, ..., -1918, -1919, -1920], dtype=int32)
- cartesian_x_vertices(vertex)float64...
- long_name :
- vertex cartesian coordinate x on unit sp
- units :
- meters
array([0.167082, 0.218918, 0.145953, ..., 0.246908, 0.224963, 0.17602 ])
- cartesian_y_vertices(vertex)float64...
- long_name :
- vertex cartesian coordinate y on unit sp
- units :
- meters
array([0.5465 , 0.585565, 0.607873, ..., 0.680831, 0.735822, 0.702503])
- cartesian_z_vertices(vertex)float64...
- long_name :
- vertex cartesian coordinate z on unit sp
- units :
- meters
array([ 0.820623, 0.780505, 0.780505, ..., -0.689569, -0.638715, -0.689569])
- edge_middle_cartesian_x(edge)float64...
- long_name :
- prime edge center cartesian coordinate x on unit sphere
- units :
- meters
array([0.156631, 0.19314 , 0.182568, ..., 0.140165, 0.114582, 0.150564])
- edge_middle_cartesian_y(edge)float64...
- long_name :
- prime edge center cartesian coordinate y on unit sphere
- units :
- meters
array([0.577607, 0.566445, 0.597154, ..., 0.711737, 0.693411, 0.684683])
- edge_middle_cartesian_z(edge)float64...
- long_name :
- prime edge center cartesian coordinate z on unit sphere
- units :
- meters
array([ 0.801147, 0.801147, 0.781074, ..., -0.68832 , -0.711373, -0.71312 ])
- edge_dual_middle_cartesian_x(edge)float64...
- long_name :
- dual edge center cartesian coordinate x on unit sphere
- units :
- meters
array([0.156324, 0.19337 , 0.182643, ..., 0.140227, 0.114253, 0.150881])
- edge_dual_middle_cartesian_y(edge)float64...
- long_name :
- dual edge center cartesian coordinate y on unit sphere
- units :
- meters
array([0.577561, 0.566235, 0.597398, ..., 0.711948, 0.69337 , 0.684444])
- edge_dual_middle_cartesian_z(edge)float64...
- long_name :
- dual edge center cartesian coordinate z on unit sphere
- units :
- meters
array([ 0.80124 , 0.80124 , 0.78087 , ..., -0.688089, -0.711466, -0.713282])
- edge_primal_normal_cartesian_x(edge)float64...
- long_name :
- unit normal to the prime edge 3D vector, coordinate x
- units :
- meters
array([-0.948047, 0.707965, 0.228364, ..., 0.195585, -0.955352, 0.739635])
- edge_primal_normal_cartesian_y(edge)float64...
- long_name :
- unit normal to the prime edge 3D vector, coordinate y
- units :
- meters
array([-0.139489, -0.645783, 0.746945, ..., 0.661582, -0.119405, -0.556637])
- edge_primal_normal_cartesian_z(edge)float64...
- long_name :
- unit normal to the prime edge 3D vector, coordinate z
- units :
- meters
array([ 0.285919, 0.285919, -0.624439, ..., 0.723917, -0.27027 , -0.378279])
- edge_dual_normal_cartesian_x(edge)float64...
- long_name :
- unit normal to the dual edge 3D vector, coordinate x
- units :
- meters
array([-0.2769 , -0.679325, 0.956305, ..., -0.970619, 0.27235 , 0.65595 ])
- edge_dual_normal_cartesian_y(edge)float64...
- long_name :
- unit normal to the dual edge 3D vector, coordinate y
- units :
- meters
array([ 0.804309, -0.511962, -0.292372, ..., 0.236093, -0.710579, 0.470493])
- edge_dual_normal_cartesian_z(edge)float64...
- long_name :
- unit normal to the dual edge 3D vector, coordinate z
- units :
- meters
array([-0.52575 , 0.52575 , -0. , ..., 0.046475, -0.64877 , 0.590225])
- cell_circumcenter_cartesian_x(cell)float64...
- long_name :
- cartesian position of the prime cell circumcenter on the unit sphere, coordinate x
- units :
- meters
array([0.177489, 0.13508 , 0.209154, ..., 0.144882, 0.092958, 0.166206])
- cell_circumcenter_cartesian_y(cell)float64...
- long_name :
- cartesian position of the prime cell circumcenter on the unit sphere, coordinate y
- units :
- meters
array([0.580541, 0.574293, 0.551646, ..., 0.727623, 0.690535, 0.672732])
- cell_circumcenter_cartesian_z(cell)float64...
- long_name :
- cartesian position of the prime cell circumcenter on the unit sphere, coordinate z
- units :
- meters
array([ 0.794651, 0.807429, 0.807429, ..., -0.670503, -0.7173 , -0.720977])
- title :
- ICON grid description
- institution :
- Max Planck Institute for Meteorology/Deutscher Wetterdienst
- source :
- git@git.mpimet.mpg.de:GridGenerator.git
- revision :
- 2cdf99bec403902989148ebb4bd38c218a64a1b3
- date :
- 20190405 at 141502
- user_name :
- Rene Redler (m300083)
- os_name :
- Linux 2.6.32-696.18.7.el6.x86_64 x86_64
- uuidOfHGrid :
- 703b3694-579c-11e9-bc94-8387e50334a2
- grid_mapping_name :
- lat_long_on_sphere
- crs_id :
- urn:ogc:def:cs:EPSG:6.0:6422
- crs_name :
- Spherical 2D Coordinate System
- ellipsoid_name :
- Sphere
- semi_major_axis :
- 6371229.0
- inverse_flattening :
- 0.0
- grid_level :
- 3
- grid_root :
- 2
- grid_ID :
- 1
- parent_grid_ID :
- 0
- no_of_subgrids :
- 1
- start_subgrid_id :
- 1
- max_childdom :
- 1
- boundary_depth_index :
- 0
- rotation_vector :
- [0. 0. 0.]
- grid_geometry :
- 1
- grid_cell_type :
- 3
- mean_edge_length :
- 480617.76530999044
- mean_dual_edge_length :
- 277650.8788228402
- mean_cell_area :
- 99629128946.83536
- mean_dual_cell_area :
- 199102708902.3395
- domain_length :
- 40031612.44147649
- domain_height :
- 40031612.44147649
- sphere_radius :
- 6371229.0
- domain_cartesian_center :
- [0. 0. 0.]
- history :
- Fri Apr 12 13:38:27 2019: ncatted -a centre,global,c,i,252 -a rotation,global,c,c,37deg around z-axis -a coverage,global,c,c,ocean only -a topography,global,c,c,modified SRTM30 -a symmetry,global,c,c,along equator -a subcentre,global,c,i,1 -a number_of_grid_used,global,c,i,30 -a ICON_grid_file_uri,global,c,c,http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0030/icon_grid_0030_R02B03_G.nc Global_IcosSymmetric_0316km_rotatedZ37d.nc icon_grid_0030_R02B03_G.nc /mnt/lustre01/work/mh0287/users/rene/GridGenerator/build/x86_64-unknown-linux-gnu/bin/grid_command
- centre :
- 252
- rotation :
- 37deg around z-axis
- coverage :
- ocean only
- topography :
- modified SRTM30
- symmetry :
- along equator
- subcentre :
- 1
- number_of_grid_used :
- 30
- ICON_grid_file_uri :
- http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0030/icon_grid_0030_R02B03_G.nc
- NCO :
- netCDF Operators version 4.7.5 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
We’ll use this little function which displays our selected grid cells on a map. It uses
voc(vertex of cell), which lists the vertices of each cell, in Fortran (1-based) index conventionclatandclon: coordinates of cell centersvlatandvlon: coordinates of vertices
The function draws a line along the cell centers in index order and colors the centers also in this order.
def show_grid(voc, clat, clon, vlat, vlon):
fig = plt.figure(figsize=(14, 7))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.Orthographic(60, 45))
ax.set_global()
ax.add_feature(cfeature.LAND, zorder=0, edgecolor="black")
plt.plot(np.rad2deg(clon), np.rad2deg(clat), transform=ccrs.Geodetic())
plt.scatter(
np.rad2deg(clon),
np.rad2deg(clat),
c=np.arange(len(clon)),
s=15,
zorder=10,
cmap="turbo",
transform=ccrs.Geodetic(),
)
for v1, v2, v3 in voc.T - 1:
i = np.array([v1, v2, v3, v1])
plt.plot(
np.rad2deg(vlon[i]),
np.rad2deg(vlat[i]),
c="k",
lw=1,
alpha=0.1,
transform=ccrs.Geodetic(),
)
plt.colorbar(label="cell index")
plt.show()
plt.close()
As we can not see anything on the backside of the globe anyways and an we know that the ICON grid is based on an icosahedron, we’ll cut out one of the faces of the icosahedron to look at the internal grid structure. We’ll call this subgrid sg1.
sg1 = grid.isel(cell=slice(2 ** (2 * 4)))
show_grid(
sg1.vertex_of_cell.values,
sg1.clat.values,
sg1.clon.values,
grid.vlat.values,
grid.vlon.values,
)
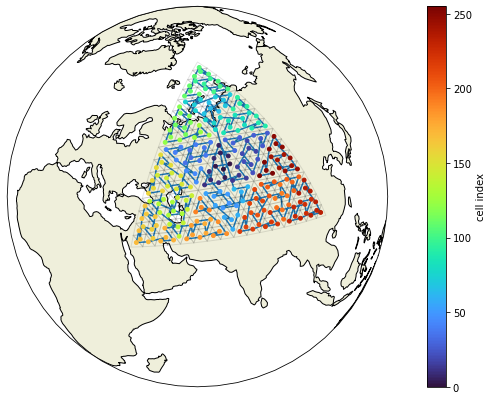
We observe that the central triangle of any larger (composed) triangle is always the first index within that group. Or the other way around: four triangles form one larger triangle and every four triangles are ordered as [center, corner1, corner2, corner3].
Let’s have a look into the vertex indices of four such connected triangles (we’ll use the first, most central triangles for the example):
sg1.vertex_of_cell.values.T[0:4]
array([[1, 2, 3],
[3, 4, 1],
[1, 5, 2],
[2, 6, 3]], dtype=int32)
Each row in this array represents one triangle and each column shows the indices of the vertices within that triangle.
We know that lower three triangles are the three corner triangles and we can see that in each triangle, the middle vertex occurs only once. As only the outer vertices of the larger triangle composed by these small triangles are not shared between inner triangles, these vertices must be the outer vertices of the larger (connected) triangle. Let’s use this information to coarsen our grid.
def coarsen_vertex_of_cell(voc):
return voc.reshape(3, -1, 4)[1, :, 1:].T
The function above reshapes the voc array into a stack of these 3x4 blocks of vertex indices and then just cuts out our three corner indices into a new, coarser voc-array.
Let’s also define a function which we can use to coarsen cell center values (e.g. clat and clon) by selecting the value of the central (first) triangle:
def coarsen_cell_center(cvalue):
return cvalue[..., ::4]
We can now apply these functions recursively to coarsen the grid several times and have a look at the resulting grid structures:
voc = sg1.vertex_of_cell.values
clat = sg1.clat.values
clon = sg1.clon.values
for i in range(3):
voc = coarsen_vertex_of_cell(voc)
clat = coarsen_cell_center(clat)
clon = coarsen_cell_center(clon)
show_grid(voc, clat, clon, grid.vlat.values, grid.vlon.values)
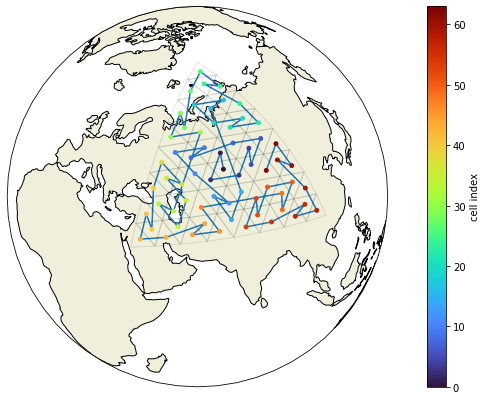
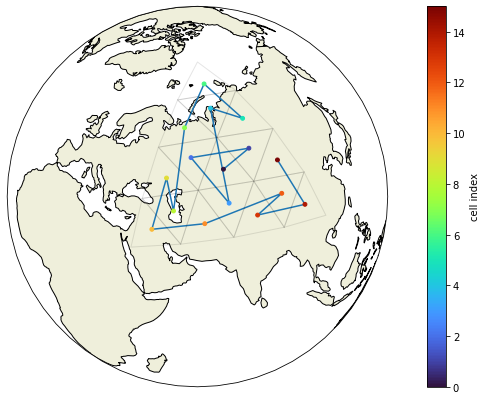
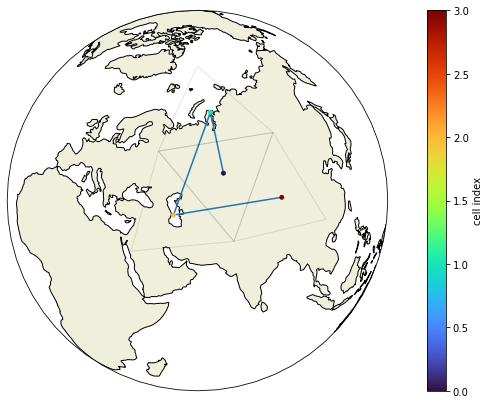
Here’s a little test which would print out the coarsening level and the cell index of any group of triangles which would violate out assumption about the ordering of the vertices. As it doesn’t print anything, we know that the method works for this grid.
voc = sg1.vertex_of_cell.values
for l in range(4):
for i, g in enumerate(voc.reshape(3, -1, 4).transpose(1, 0, 2)):
if np.any(g[:, 0, np.newaxis] == g[1, 1:]):
print(l, i, g)
voc = coarsen_vertex_of_cell(voc)
We can also apply the coarsening several times to our full grid to make a little image of the grid structure on the globe:
voc = coarsen_vertex_of_cell(coarsen_vertex_of_cell(grid.vertex_of_cell.values))
fig = plt.figure(figsize=(14, 7))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.Orthographic(60, 45))
ax.set_global()
ax.add_feature(cfeature.LAND, zorder=0, edgecolor="black")
for v1, v2, v3 in voc.T - 1:
i = np.array([v1, v2, v3, v1])
plt.plot(
np.rad2deg(grid.vlon.values[i]),
np.rad2deg(grid.vlat.values[i]),
c="k",
lw=1,
alpha=0.1,
transform=ccrs.Geodetic(),
)
plt.show()
plt.close()