Limited-area HEALPix#
The HEALPix grid is designed to cover an entire sphere, be it Space or the Earth. This raises the question of whether HEALPix can also be used to store regional cutouts, as is often the case in weather forecasting or regional climate modelling.
In this article we will try to outline some technical issues and suggest ways around them.
import cartopy.crs as ccrs
import easygems.healpix as egh
import healpix as hp
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
Define two helper functions to create a cartopy map with regional extent and plot boxes on it.
def get_axis_europe():
"""Create cartyop.GeoAxis with an extent centered around Europe."""
_, ax = plt.subplots(subplot_kw={"projection": ccrs.PlateCarree()})
ax.coastlines()
ax.set_extent([-40, 60, 20, 65])
return ax
def plot_extent_box(ax, extent, edgecolor="m", linewidth=2):
"""Add a rectangular patch around an extent to a cartopy.GeoAxis."""
return ax.add_patch(
plt.Rectangle(
xy=[extent[0], extent[2]],
width=extent[1] - extent[0],
height=extent[3] - extent[2],
facecolor="none",
edgecolor=edgecolor,
linewidth=linewidth,
transform=ccrs.PlateCarree(),
)
)
Querying grid points#
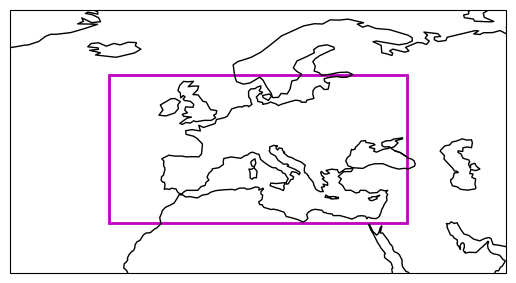
In a first step, we will define a simple lat/lon box over Europe as our region of interest.
extent = [-20, 40, 30, 60] # Europe
ax = get_axis_europe()
plot_extent_box(ax, extent);
Next, we can use existing HEALPix libraries to compute lat/lon coordinates at a given order (zoom level). These coordinates can then be compared again our boundaries to query a regional cutout.
# Compute HEALPix lat/lon coordinates for a given order (zoom level)
order = zoom = 8
nside = hp.order2nside(order)
npix = hp.nside2npix(nside)
hp_lon, hp_lat = hp.pix2ang(nside, np.arange(npix), nest=True, lonlat=True)
hp_lon = (hp_lon + 180) % 360 - 180
# Find all grid points within the defined extent
icell, = np.where(
(hp_lon > extent[0]) &
(hp_lon < extent[1]) &
(hp_lat > extent[2]) &
(hp_lat < extent[3])
)
# Plot the selected points
ax = get_axis_europe()
plot_extent_box(ax, extent)
region = np.full_like(hp_lon, fill_value=np.nan) # Full HEALPix with NaNs
region[icell] = 1 # Set selected values to 1
egh.healpix_show(region);
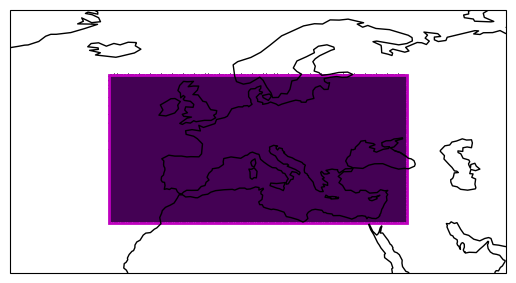
It worked! However, a technical problem with these slices is that they distort the grid index ordering[1].
Maintaining the grid index ordering#
The nested HEALPix grid ordering ensures that geographical proximity of data is mapped to proximity in the array. However, this relationship is broken when we cut along simple lines on the map.
Fortunately, we can maintain the proximity by keeping full chunks that follow the \(4^n\) relation.
We will use the get_full_chunks() convenience function provided by easygems.
This function takes an array of indices and a chunk size and returns all indices of grid points in chunks that are touched.
# Get indices of **full chunks**
ichunk = egh.get_full_chunks(icell, chunksize=4**5)
# Plot the selected points
ax = get_axis_europe()
plot_extent_box(ax, extent)
region = np.full_like(hp_lon, fill_value=np.nan) # Full HEALPix with NaNs
region[ichunk] = 1 # Set selected values to 1
egh.healpix_show(region);
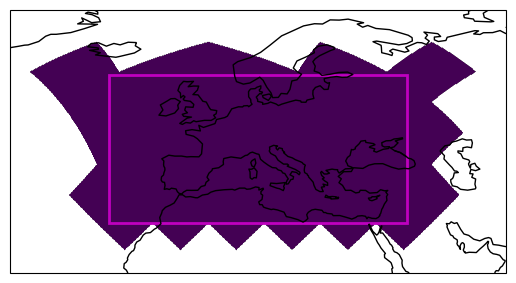
Setting the Coordinate References System#
An open question is what to store in the Coordinate Reference System (CRS). Our suggestion is to store the information about the full global HEALPix grid. This way downstream libraries will be able to compute the full grid.
crs = xr.DataArray(
name="crs",
attrs={
"grid_mapping_name": "healpix",
"healpix_nside": 2**zoom,
"healpix_order": "nest",
},
)
crs
<xarray.DataArray 'crs' ()> Size: 8B
array(nan)
Attributes:
grid_mapping_name: healpix
healpix_nside: 256
healpix_order: nestIn addition, the arrays in the dataset must contain the global indices of the original HEALPix grid.
This allows the original location of any selected grid point to be diagnosed.
You can set the values of indices outside the defined extent (not inside icell) to NaN, but the index must remain to keep the array order in tact.
Here is a complete example of a limited-area HEALPix dataset:
# Construct global HEALPix dataset with proper `cell` **coordinate** and CRS information.
ds = xr.Dataset(
coords={
"crs": crs,
"lat": (("cell",), hp_lat),
"lon": (("cell",), hp_lon),
"cell": (("cell",), np.arange(npix)),
}
)
# Select the limited-area while maintaining global index information (and chunks)
ds.isel(cell=ichunk)
<xarray.Dataset> Size: 860kB
Dimensions: (cell: 35840)
Coordinates:
* cell (cell) int64 287kB 14336 14337 14338 14339 ... 327677 327678 327679
crs float64 8B nan
lat (cell) float64 287kB 24.79 24.95 24.95 25.12 ... 41.41 41.41 41.61
lon (cell) float64 287kB 39.38 39.55 39.2 39.38 ... 0.1758 -0.1758 0.0
Data variables:
*empty*