The km-scale cloud by DKRZ#
The km-scale cloud formerly known as EERIE cloud addresses data users who cannot work on DKRZ´s High Performance Computer Levante next to the data. It provides open, efficient, Zarr-based access to High-resolution ESM datasets produced by
projects like EERIE and NextGEMs
models like ICON and IFS-FESOM.
Whether originally stored as Zarr, NetCDF, or GRIB, the km-scale cloud delivers - without rewriting, relocating, or converting - byte ranges of the original files as Zarr chunks. In addition, it incorporates Dask for on-demand, server-side data reduction tasks — such as compression and the computation of summary statistics. This combination makes it practical and scalable to work with petabyte-scale climate data even to users with limited compute resources. Integrated support for catalog tools like STAC (SpatioTemporal Asset Catalog) and intake, organized in a semantic hierarchy, allows users to discover, query, and subset data efficiently through standardized, interoperable interfaces.
This is enabled by the python package cloudify described in detail in Wachsmann, 2025.
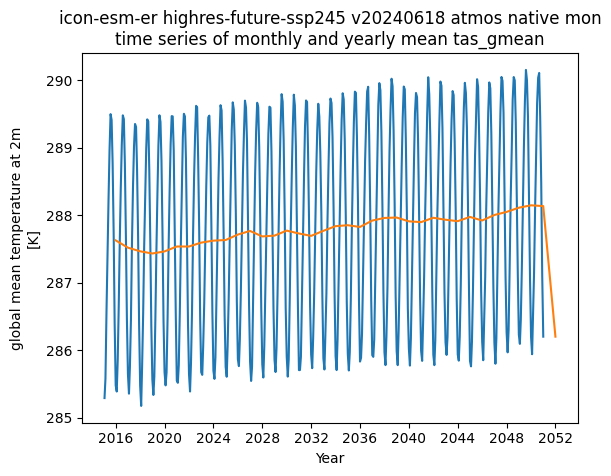
Show case: Time series quickplot for a ICON simulation#
import xarray as xr
dataset_id="icon-esm-er.highres-future-ssp245.v20240618.atmos.native.mon"
ds=xr.open_dataset(
f"https://km-scale-cloud.dkrz.de/datasets/{dataset_id}/kerchunk",
engine="zarr",
chunks="auto",
zarr_format=2,
)
import matplotlib.pyplot as plt
var="tas_gmean"
vmean = ds[var].squeeze()
vmean_yr = vmean.resample(time="YE").mean()
vmean.plot()
vmean_yr.plot()
plt.title(" ".join(dataset_id.split(".")) + f"\ntime series of monthly and yearly mean {var}")
plt.xlabel("Year")
Text(0.5, 0, 'Year')
Km-scale datasets and how to approach them#
The km-scale-cloud provides many endpoints (URIs) with different features. Under this link, all available endpoints are documented.
Helpful dataset endpoints#
A key endpoint is the /datasets endpoint which provides a simple list of datasets accessible through the km-scale-cloud. We can use it like:
import requests
kmscale_uri = "https://km-scale-cloud.dkrz.de"
kmscale_datasets_uri=kmscale_uri+"/datasets"
kmscale_datasets=requests.get(kmscale_datasets_uri).json()
print(f"The km-scale-cloud provides {len(kmscale_datasets)} datasests such as:")
print(kmscale_datasets[0])
The km-scale-cloud provides 571 datasests such as:
cosmo-rea-1hrPt_atmos
For each dataset, a Xarray-dataset view endpoint exists which can be helpful to get an overview about the content of the dataset. The endpoint is constructed like datasets/dataset_id/, e.g.:
dataset_pattern="atmos.native.mon"
global_atm_mon_mean_ds_name=next(
a for a in kmscale_datasets if dataset_pattern in a
)
print(
"An available atmospheric global mean monthly mean dataset is: "+
global_atm_mon_mean_ds_name
)
global_atm_mon_mean_ds_uri=kmscale_datasets_uri+"/"+global_atm_mon_mean_ds_name
An available atmospheric global mean monthly mean dataset is: icon-epoc.control-1990.v20250325.atmos.native.mon
The key endpoint is the default Zarr-endpoint constructed like datasets/DATASET_ID/kerchunk. Such an address can be used by any tool that can read Zarr over http. E.g. With Xarray:
xr.open_dataset(
global_atm_mon_mean_ds_uri+"/kerchunk",
engine="zarr"
)
<xarray.Dataset> Size: 30kB
Dimensions: (time: 420, lat: 1, lon: 1)
Coordinates:
* time (time) datetime64[ns] 3kB 1990-02-01 ... 2025-01-01
* lat (lat) float64 8B 0.0
* lon (lon) float64 8B 0.0
Data variables: (12/16)
duphyvi_gmean (time, lat, lon) float32 2kB ...
evap_gmean (time, lat, lon) float32 2kB ...
fwfoce_gmean (time, lat, lon) float32 2kB ...
kedisp_gmean (time, lat, lon) float32 2kB ...
prec_gmean (time, lat, lon) float32 2kB ...
radbal_gmean (time, lat, lon) float32 2kB ...
... ...
tas_gmean (time, lat, lon) float32 2kB ...
udynvi_gmean (time, lat, lon) float32 2kB ...
ufcs_gmean (time, lat, lon) float32 2kB ...
ufts_gmean (time, lat, lon) float32 2kB ...
ufvs_gmean (time, lat, lon) float32 2kB ...
uphybal_gmean (time, lat, lon) float32 2kB ...
Attributes:
CDI: Climate Data Interface version 2.4.0 (https://mpimet.mpg.de...
Conventions: CF-1.6
comment: Helmuth Haak (shared) (b383127) on l30288 (Linux 4.18.0-477...
history: ./icon at 20241025 152041
institution: Max Planck Institute for Meteorology/Deutscher Wetterdienst
references: see MPIM/DWD publications
source: version: 2024.10; revision: icon-2024.10-5-g59df2b1543c98c8...
title: ICON simulation- time: 420
- lat: 1
- lon: 1
- time(time)datetime64[ns]1990-02-01 ... 2025-01-01
- axis :
- T
- standard_name :
- time
array(['1990-02-01T00:00:00.000000000', '1990-03-01T00:00:00.000000000', '1990-04-01T00:00:00.000000000', ..., '2024-11-01T00:00:00.000000000', '2024-12-01T00:00:00.000000000', '2025-01-01T00:00:00.000000000'], shape=(420,), dtype='datetime64[ns]') - lat(lat)float640.0
- axis :
- Y
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
array([0.])
- lon(lon)float640.0
- axis :
- X
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([0.])
- duphyvi_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean vertically integrated moist internal energy change by physics
- standard_name :
- duphyvi_gmean
- units :
- J m-2
[420 values with dtype=float32]
- evap_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean evaporation flux
- standard_name :
- evap_gmean
- units :
- kg m-2 s-1
[420 values with dtype=float32]
- fwfoce_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean surface freshwater flux over ocean surface
- standard_name :
- fwfoce_gmean
- units :
- kg m-2 s-1
[420 values with dtype=float32]
- kedisp_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean vert. integr. dissip. kin. energy
- standard_name :
- kedisp_gmean
- units :
- W m-2
[420 values with dtype=float32]
- prec_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean precipitation flux
- standard_name :
- prec_gmean
- units :
- kg m-2 s-1
[420 values with dtype=float32]
- radbal_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean net radiative flux into atmosphere
- standard_name :
- radbal_gmean
- units :
- W m-2
[420 values with dtype=float32]
- radtop_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean toa net total radiation
- standard_name :
- radtop_gmean
- units :
- W m-2
[420 values with dtype=float32]
- rlut_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean toa outgoing longwave radiation
- standard_name :
- rlut_gmean
- units :
- W m-2
[420 values with dtype=float32]
- rsdt_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean toa incident shortwave radiation
- standard_name :
- rsdt_gmean
- units :
- W m-2
[420 values with dtype=float32]
- rsut_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean toa outgoing shortwave radiation
- standard_name :
- rsut_gmean
- units :
- W m-2
[420 values with dtype=float32]
- tas_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- global mean temperature at 2m
- standard_name :
- tas_gmean
- units :
- K
[420 values with dtype=float32]
- udynvi_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean vertically integrated moist internal energy after dynamics
- standard_name :
- udynvi_gmean
- units :
- J m-2
[420 values with dtype=float32]
- ufcs_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean energy flux at surface from condensate
- standard_name :
- ufcs_gmean
- units :
- W m-2
[420 values with dtype=float32]
- ufts_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean energy flux at surface from thermal exchange
- standard_name :
- ufts_gmean
- units :
- W m-2
[420 values with dtype=float32]
- ufvs_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean energy flux at surface from vapor exchange
- standard_name :
- ufvs_gmean
- units :
- W m-2
[420 values with dtype=float32]
- uphybal_gmean(time, lat, lon)float32...
- code :
- 255
- long_name :
- mean energy balance in aes physics
- standard_name :
- uphybal_gmean
- units :
- W m-2
[420 values with dtype=float32]
- CDI :
- Climate Data Interface version 2.4.0 (https://mpimet.mpg.de/cdi)
- Conventions :
- CF-1.6
- comment :
- Helmuth Haak (shared) (b383127) on l30288 (Linux 4.18.0-477.58.1.el8_8.x86_64 x86_64)
- history :
- ./icon at 20241025 152041
- institution :
- Max Planck Institute for Meteorology/Deutscher Wetterdienst
- references :
- see MPIM/DWD publications
- source :
- version: 2024.10; revision: icon-2024.10-5-g59df2b1543c98c80fbd96124c3f9b587a4680d19-dirty; URL: git@gitlab.dkrz.de:icon/icon-mpim.git
- title :
- ICON simulation
The Datatree: All in one Zarr-group#
Note: xarray>=2025.7.1 required and evolving fast.
Opening Zarr and representing it in Xarray does not coast much resources. It is fast and small in memory. A strategy to work with multiple datasets is therefore to just open everything.
We can access to the full >10PB of the km-scale-cloud with only using xarray by applying the following code:
dt = xr.open_datatree(
"https://km-scale-cloud.dkrz.de/datasets",
engine="zarr",
zarr_format=2,
chunks=None,
create_default_indexes=False,
decode_cf=False,
)
The dt object is a Xarray Datatree. This datatree object allows you to browse and discover the full content of the km-scale-cloud similar to the functionality of intake but without requiring an additional tool.
The workflow using the datatree involves the following steps:
filter: subset the tree so that it only includes the datasets you want. You can apply any customized function using the datasets as input. In the following example, we look for a specific dataset name.chunk,decode_cf: in case you need coordinate values for subsetting or in case you are interested in more than one chunk, better use these functions to allow lazy access on coordinates.Instead of writing loops over a list or a dictionary of datasets, you can now use the
.map_over_datasetsfunction to apply a function to all datasets at once.
path_filter="s2024-08-10"
filtered_tree=dt.filter(lambda ds: ds if path_filter in ds.path else None)
display(filtered_tree)
<xarray.DataTree> Group: /
filtered_tree_chunked=filtered_tree.chunk()
filtered_tree_chunked_decoded=filtered_tree_chunked.map_over_datasets(
lambda ds: xr.decode_cf(ds) if "time" in ds else None
)
Any .compute() or .load() will trigger data retrieval. Thus, make sure you first subset before you download.
The .nbytes object shows you how much uncompressed data will be loaded to your memory.
filtered_tree_chunked_decoded.nbytes/1024**2
0.0
time_sel="2024-08-10"
var="tas"
filtered_tree_chunked_decoded_subsetted=filtered_tree_chunked_decoded.map_over_datasets(
lambda ds: ds[[var]].sel(time=time_sel) if all(a in ds for a in [var,"time"]) else None
).prune()
print(filtered_tree_chunked_decoded_subsetted.nbytes/1024**2)
0.0
Before running .load(), add all lazy functions to the dask´s taskgraph:
dt_workflow=filtered_tree_chunked_decoded_subsetted.map_over_datasets(lambda ds: ds.mean(dim="time") if "time" in ds else None)
%%time
ds_mean=dt_workflow.compute().leaves[0].to_dataset()
ds_mean
CPU times: user 407 μs, sys: 0 ns, total: 407 μs
Wall time: 439 μs
<xarray.Dataset> Size: 0B
Dimensions: ()
Data variables:
*empty*STAC interface#
The most interoperable catalog interface for the km-scale-cloud is with STAC (SpatioTemporal Asset Catalogs). You can use any stac-browser implementation to browse and discover the dynamically created km-scale cloud collection with all datasets linked as dynamic items.
For programmatic access, follow this notebook.
import pystac
kmscale_collection_uri="https://km-scale-cloud.dkrz.de/stac-collection-all.json"
kmscale_collection=pystac.Collection.from_file(kmscale_collection_uri)
kmscale_collection
- type "Collection"
- id "eerie-cloud-all"
- stac_version "1.1.0"
- description " # Items of the eerie.cloud DKRZ hosts a data server named ‘eerie.cloud’ for global high resolution Earth System Model simulation output stored at the German Climate Computing Center (DKRZ). This was developped within the EU project EERIE. Eerie.cloud makes use of the python package xpublish. Xpublish is a plugin for xarray (Hoyer, 2023) which is widely used in the Earth System Science community. It serves ESM output formatted as zarr (Miles, 2020) via a RestAPI based on FastAPI. Served in this way, the data imitates cloud-native data (Abernathey, 2021) and features many capabilities of cloud-optimized data. [Imprint](https://www.dkrz.de/en/about-en/contact/impressum) and [Privacy Policy](https://www.dkrz.de/en/about-en/contact/en-datenschutzhinweise). "
links[] 574 items
0
- rel "self"
- href "https://km-scale-cloud.dkrz.de/stac-collection-all.json"
- type "application/json"
1
- rel "root"
- href "https://eerie.cloud.dkrz.de/stac-collection-all.json"
- type "application/json"
- title "ESM data from DKRZ in Zarr format"
2
- rel "parent"
- href "https://swift.dkrz.de/v1/dkrz_7fa6baba-db43-4d12-a295-8e3ebb1a01ed/catalogs/stac-catalog-eeriecloud.json"
- type "application/json"
3
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/stac"
- type "application/json"
- title "cosmo-rea-1hrPt_atmos"
4
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_land/stac"
- type "application/json"
- title "cosmo-rea-1hrPt_land"
5
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_landIce/stac"
- type "application/json"
- title "cosmo-rea-1hrPt_landIce"
6
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hr_atmos/stac"
- type "application/json"
- title "cosmo-rea-1hr_atmos"
7
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hr_land/stac"
- type "application/json"
- title "cosmo-rea-1hr_land"
8
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-6hr_atmos/stac"
- type "application/json"
- title "cosmo-rea-6hr_atmos"
9
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-day_atmos/stac"
- type "application/json"
- title "cosmo-rea-day_atmos"
10
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-day_land/stac"
- type "application/json"
- title "cosmo-rea-day_land"
11
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-day_landIce/stac"
- type "application/json"
- title "cosmo-rea-day_landIce"
12
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-mon_atmos/stac"
- type "application/json"
- title "cosmo-rea-mon_atmos"
13
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-mon_land/stac"
- type "application/json"
- title "cosmo-rea-mon_land"
14
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-mon_landIce/stac"
- type "application/json"
- title "cosmo-rea-mon_landIce"
15
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_daily_healpix128_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_daily_healpix128_ocean"
16
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_daily_healpix512_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_daily_healpix512_ocean"
17
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_hourly_0.25deg/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_hourly_0.25deg"
18
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_hourly_healpix128/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_hourly_healpix128"
19
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_hourly_healpix2048/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_hourly_healpix2048"
20
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_monthly_0.25deg/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_monthly_0.25deg"
21
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_monthly_healpix128/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_monthly_healpix128"
22
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.2D_monthly_healpix2048/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.2D_monthly_healpix2048"
23
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_daily_healpix128_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_daily_healpix128_ocean"
24
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_daily_healpix512_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_daily_healpix512_ocean"
25
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_hourly_0.25deg/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_hourly_0.25deg"
26
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_hourly_0.25deg_snow/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_hourly_0.25deg_snow"
27
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix128/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix128"
28
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix128_snow/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix128_snow"
29
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix2048/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix2048"
30
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix2048_snow/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production.3D_hourly_healpix2048_snow"
31
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_daily_healpix128_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_daily_healpix128_ocean"
32
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_daily_healpix512_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_daily_healpix512_ocean"
33
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_hourly_0.25deg/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_hourly_0.25deg"
34
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_hourly_healpix128/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_hourly_healpix128"
35
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_hourly_healpix2048/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_hourly_healpix2048"
36
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_monthly_0.25deg/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_monthly_0.25deg"
37
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_monthly_healpix128/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_monthly_healpix128"
38
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_monthly_healpix2048/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.2D_monthly_healpix2048"
39
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_daily_healpix128_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_daily_healpix128_ocean"
40
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_daily_healpix512_ocean/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_daily_healpix512_ocean"
41
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_0.25deg/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_0.25deg"
42
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_0.25deg_snow/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_0.25deg_snow"
43
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix128/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix128"
44
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix128_snow/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix128_snow"
45
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix2048/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix2048"
46
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix2048_snow/stac"
- type "application/json"
- title "nextgems.IFS_2.8-FESOM_5-production-deep-off.3D_hourly_healpix2048_snow"
47
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/orcestra_1250m_2d_hpz12/stac"
- type "application/json"
- title "orcestra_1250m_2d_hpz12"
48
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/orcestra_1250m_3d_hpz12/stac"
- type "application/json"
- title "orcestra_1250m_3d_hpz12"
49
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.pressure-level_analysis_daily/stac"
- type "application/json"
- title "era5-dkrz.pressure-level_analysis_daily"
50
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.pressure-level_analysis_monthly/stac"
- type "application/json"
- title "era5-dkrz.pressure-level_analysis_monthly"
51
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.surface_analysis_daily/stac"
- type "application/json"
- title "era5-dkrz.surface_analysis_daily"
52
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.surface_analysis_hourly/stac"
- type "application/json"
- title "era5-dkrz.surface_analysis_hourly"
53
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.surface_analysis_monthly/stac"
- type "application/json"
- title "era5-dkrz.surface_analysis_monthly"
54
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.surface_forecast_hourly/stac"
- type "application/json"
- title "era5-dkrz.surface_forecast_hourly"
55
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/era5-dkrz.surface_forecast_monthly/stac"
- type "application/json"
- title "era5-dkrz.surface_forecast_monthly"
56
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
57
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
58
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
59
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
60
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
61
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
62
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
63
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
64
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
65
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
66
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
67
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.dayPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.dayPt"
68
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fxPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fxPt"
69
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
70
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.monPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.monPt"
71
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.MEU-3.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
72
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon"
73
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt"
74
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day"
75
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt"
76
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp585.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx"
77
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon"
78
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt"
79
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day"
80
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt"
81
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp245.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx"
82
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon"
83
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt"
84
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day"
85
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt"
86
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx"
87
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hr"
88
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.historical.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hr"
89
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon"
90
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt"
91
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day"
92
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt"
93
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx"
94
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hr"
95
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp126.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hr"
96
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.mon"
97
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hrPt"
98
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.day"
99
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hrPt"
100
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.fx"
101
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.1hr"
102
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-BTU.CNRM-ESM2-1.ssp370.r1i1p1f2.ICON-CLM-202407-1-1.v1-r1.6hr"
103
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
104
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
105
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
106
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
107
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
108
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
109
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
110
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
111
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
112
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
113
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
114
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
115
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
116
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-GERICS.EC-Earth3-Veg.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
117
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.mon"
118
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.6hrPt"
119
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.day"
120
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.1hrPt"
121
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.fx"
122
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.1hr"
123
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r2.6hr"
124
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
125
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
126
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
127
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
128
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
129
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
130
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.ERA5.evaluation.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
131
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
132
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
133
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
134
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
135
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
136
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
137
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-Hereon.EC-Earth3-Veg.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
138
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
139
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
140
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
141
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
142
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
143
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp585.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
144
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
145
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
146
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
147
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
148
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
149
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
150
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
151
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
152
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
153
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
154
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
155
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
156
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
157
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
158
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
159
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
160
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
161
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
162
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
163
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
164
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
165
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
166
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
167
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-DWD.MPI-ESM1-2-HR.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
168
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
169
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp245.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
170
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
171
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
172
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
173
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
174
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
175
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
176
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.historical.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
177
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
178
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
179
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
180
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
181
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
182
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
183
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp126.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
184
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.mon"
185
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hrPt"
186
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.day"
187
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hrPt"
188
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.fx"
189
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.1hr"
190
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr/stac"
- type "application/json"
- title "cordex-cmip6.DD.EUR-12.CLMcom-KIT.MIROC6.ssp370.r1i1p1f1.ICON-CLM-202407-1-1.v1-r1.6hr"
191
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.2d_6h_inst/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.2d_6h_inst"
192
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.2d_daily_max/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.2d_daily_max"
193
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.2d_daily_mean"
194
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.2d_daily_min/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.2d_daily_min"
195
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.2d_monthly_mean"
196
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.mon/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.mon"
197
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.atmos.native.pl_6h_inst/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.atmos.native.pl_6h_inst"
198
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.land.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.land.native.2d_daily_mean"
199
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.land.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.land.native.2d_monthly_mean"
200
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.land.native.mon/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.land.native.mon"
201
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.2d_daily_mean"
202
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.2d_monthly_mean"
203
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.2d_monthly_square/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.2d_monthly_square"
204
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.age_monthly_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.age_monthly_mean"
205
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.eddy_monthly_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.eddy_monthly_mean"
206
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.layers_monthly_mean/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.layers_monthly_mean"
207
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-epoc.control-1990.v20250325.ocean.native.mon/stac"
- type "application/json"
- title "icon-epoc.control-1990.v20250325.ocean.native.mon"
208
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/tape_test/stac"
- type "application/json"
- title "tape_test"
209
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.gr025.daily/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.gr025.daily"
210
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_daily_center/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_daily_center"
211
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_daily_edge/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_daily_edge"
212
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_aermon/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_aermon"
213
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_amon_center/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_amon_center"
214
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_amon_edge/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_amon_edge"
215
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_emon/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.atmos.native.atmos_monthly_emon"
216
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.ocean.gr025.daily/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.ocean.gr025.daily"
217
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n216-orca025.eerie-picontrol.ocean.gr025.monthly/stac"
- type "application/json"
- title "hadgem3-gc5-n216-orca025.eerie-picontrol.ocean.gr025.monthly"
218
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.gr025.daily/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.gr025.daily"
219
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_daily_center/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_daily_center"
220
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_daily_edge/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_daily_edge"
221
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_aermon/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_aermon"
222
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_amon_center/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_amon_center"
223
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_amon_edge/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_amon_edge"
224
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_emon/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.atmos.native.atmos_monthly_emon"
225
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.ocean.gr025.daily/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.ocean.gr025.daily"
226
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/hadgem3-gc5-n640-orca12.eerie-picontrol.ocean.gr025.monthly/stac"
- type "application/json"
- title "hadgem3-gc5-n640-orca12.eerie-picontrol.ocean.gr025.monthly"
227
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_daily_max/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_daily_max"
228
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_daily_mean"
229
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_daily_min/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_daily_min"
230
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.2d_monthly_mean"
231
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.pfull_phalf_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.pfull_phalf_monthly_mean"
232
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.plev19_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.plev19_daily_mean"
233
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.plev19_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.atmos.gr025.plev19_monthly_mean"
234
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_mean"
235
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_mean_sst_50year/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_mean_sst_50year"
236
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_mean_vertical"
237
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_square/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_daily_square"
238
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_monthly_mean"
239
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_monthly_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_monthly_mean_vertical"
240
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_monthly_square/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.2d_monthly_square"
241
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.5lev_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.5lev_daily_mean"
242
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.5lev_daily_mean_w/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.5lev_daily_mean_w"
243
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.eddy_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.eddy_monthly_mean"
244
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.ml_1mth_mean/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.ml_1mth_mean"
245
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.ml_1mth_mean_w/stac"
- type "application/json"
- title "icon-esm-er.eerie-control-1950.v20240618.ocean.gr025.ml_1mth_mean_w"
246
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_6hr_inst/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_6hr_inst"
247
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_6hr_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_6hr_mean"
248
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_daily_max/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_daily_max"
249
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_daily_mean"
250
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_daily_min/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_daily_min"
251
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.2d_monthly_mean"
252
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.pfull_1d_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.pfull_1d_mean"
253
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.pfull_phalf_1mth_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.pfull_phalf_1mth_mean"
254
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.pl_6hr_inst/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.pl_6hr_inst"
255
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.plev19_1d_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.plev19_1d_mean"
256
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.plev19_1mth_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.gr025.plev19_1mth_mean"
257
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_1h_inst/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_1h_inst"
258
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_1h_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_1h_mean"
259
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_3h_inst/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_3h_inst"
260
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_6h_inst/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_6h_inst"
261
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_6h_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_6h_mean"
262
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_daily_max/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_daily_max"
263
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_daily_mean"
264
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_daily_min/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_daily_min"
265
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.2d_monthly_mean"
266
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.model-level_daily_mean_1/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.model-level_daily_mean_1"
267
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.model-level_daily_mean_2/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.model-level_daily_mean_2"
268
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.model-level_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.model-level_monthly_mean"
269
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.mon/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.mon"
270
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.atmos.native.pl_6h_inst/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.atmos.native.pl_6h_inst"
271
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.land.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.land.native.2d_daily_mean"
272
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.land.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.land.native.2d_monthly_mean"
273
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.land.native.mon/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.land.native.mon"
274
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_daily_mean"
275
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_daily_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_daily_mean_vertical"
276
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_daily_square/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_daily_square"
277
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_monthly_mean"
278
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_monthly_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_monthly_mean_vertical"
279
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_monthly_square/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.2d_monthly_square"
280
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.5lev_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.5lev_daily_mean"
281
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.5lev_daily_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.5lev_daily_mean_vertical"
282
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.model-level_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.model-level_monthly_mean"
283
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.model-level_monthly_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.gr025.model-level_monthly_mean_vertical"
284
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_daily_mean"
285
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_daily_square/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_daily_square"
286
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_monthly_mean"
287
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_monthly_square/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.2d_monthly_square"
288
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.5lev_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.5lev_daily_mean"
289
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.eddy_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.eddy_monthly_mean"
290
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.moc_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.moc_monthly_mean"
291
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.model-level_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.model-level_daily_mean"
292
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.model-level_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.model-level_monthly_mean"
293
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.highres-future-ssp245.v20240618.ocean.native.mon/stac"
- type "application/json"
- title "icon-esm-er.highres-future-ssp245.v20240618.ocean.native.mon"
294
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_6hourly_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_6hourly_inst"
295
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_6hourly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_6hourly_mean"
296
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_daily_max/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_daily_max"
297
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_daily_mean"
298
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_daily_min/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_daily_min"
299
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_hourly_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_hourly_inst"
300
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_hourly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_hourly_mean"
301
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_monthly_mean"
302
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.pfull_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.pfull_daily_mean"
303
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.pfull_phalf_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.pfull_phalf_monthly_mean"
304
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.pl_6hourly_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.pl_6hourly_inst"
305
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.plev19_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.plev19_daily_mean"
306
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.plev19_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.gr025.plev19_monthly_mean"
307
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_1h_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_1h_inst"
308
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_1h_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_1h_mean"
309
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_3h_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_3h_inst"
310
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_6h_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_6h_inst"
311
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_6h_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_6h_mean"
312
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_daily_max/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_daily_max"
313
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_daily_mean"
314
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_daily_min/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_daily_min"
315
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.2d_monthly_mean"
316
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.model-level_daily_mean_1/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.model-level_daily_mean_1"
317
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.model-level_daily_mean_2/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.model-level_daily_mean_2"
318
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.model-level_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.model-level_monthly_mean"
319
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.mon/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.mon"
320
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.native.pl_6h_inst/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.atmos.native.pl_6h_inst"
321
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.land.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.land.native.2d_daily_mean"
322
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.land.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.land.native.2d_monthly_mean"
323
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.land.native.mon/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.land.native.mon"
324
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_1d_mean_ssh/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_1d_mean_ssh"
325
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_daily_mean"
326
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_daily_square/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_daily_square"
327
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_monthly_mean"
328
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_monthly_mean_vertical/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_monthly_mean_vertical"
329
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_monthly_square/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.2d_monthly_square"
330
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.5lev_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.5lev_daily_mean"
331
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.5lev_daily_w_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.5lev_daily_w_mean"
332
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.eddy_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.eddy_monthly_mean"
333
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.ml_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.ml_monthly_mean"
334
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.gr025.ml_monthly_w_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.gr025.ml_monthly_w_mean"
335
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.2d_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.2d_daily_mean"
336
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.2d_daily_square/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.2d_daily_square"
337
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.2d_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.2d_monthly_mean"
338
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.2d_monthly_square/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.2d_monthly_square"
339
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.5lev_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.5lev_daily_mean"
340
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.eddy_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.eddy_monthly_mean"
341
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.moc_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.moc_monthly_mean"
342
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.model-level_daily_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.model-level_daily_mean"
343
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.model-level_monthly_mean/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.model-level_monthly_mean"
344
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.ocean.native.mon/stac"
- type "application/json"
- title "icon-esm-er.hist-1950.v20240618.ocean.native.mon"
345
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_24h"
346
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h"
347
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h_acc"
348
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_monthly"
349
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_24h"
350
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_6h"
351
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_monthly"
352
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h"
353
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h_wam/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h_wam"
354
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h"
355
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_acc"
356
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_wam/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_wam"
357
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly"
358
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly_wam/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly_wam"
359
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.3D_24h"
360
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.3D_6h"
361
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.atmos.native.3D_monthly"
362
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.wave.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist-c-0-a-lr20.v20240901.wave.gr025.2D_monthly"
363
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_24h"
364
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_6h"
365
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_6h_acc"
366
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.2D_monthly"
367
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.3D_24h"
368
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.3D_6h"
369
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.gr025.3D_monthly"
370
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_24h"
371
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_24h_wam/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_24h_wam"
372
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_6h"
373
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_6h_acc"
374
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_6h_wam/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_6h_wam"
375
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_monthly"
376
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.2D_monthly_wam/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.2D_monthly_wam"
377
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.3D_24h"
378
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.3D_6h"
379
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.atmos.native.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.atmos.native.3D_monthly"
380
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco1279.hist.v20240901.wave.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco1279.hist.v20240901.wave.gr025.2D_monthly"
381
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_1h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_1h"
382
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_1h_acc/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_1h_acc"
383
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_24h"
384
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.2D_monthly"
385
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.3D_1h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.3D_1h"
386
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.3D_24h"
387
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20240901.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20240901.atmos.gr025.3D_monthly"
388
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h"
389
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h_acc/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h_acc"
390
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h_acc_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h_acc_extra"
391
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_1h_extra"
392
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_24h"
393
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_24h_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_24h_extra"
394
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_monthly"
395
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_monthly_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.2D_monthly_extra"
396
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.3D_1h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.3D_1h"
397
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.3D_24h"
398
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.gr025.3D_monthly"
399
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h"
400
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h_acc/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h_acc"
401
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h_acc_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h_acc_extra"
402
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.2D_1h_extra"
403
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.2D_24h"
404
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.2D_24h_extra/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.2D_24h_extra"
405
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.3D_1h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.3D_1h"
406
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco2559.hist.v20250101.atmos.native.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco2559.hist.v20250101.atmos.native.3D_24h"
407
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_24h"
408
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h"
409
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_6h_acc"
410
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.2D_monthly"
411
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_24h"
412
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_6h"
413
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.gr025.3D_monthly"
414
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h"
415
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_24h_wam"
416
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h"
417
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_acc"
418
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_6h_wam"
419
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly"
420
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.2D_monthly_wam"
421
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.3D_24h"
422
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.3D_6h"
423
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.atmos.native.3D_monthly"
424
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-0-a-lr20.v20240901.wave.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-0-a-lr20.v20240901.wave.gr025.2D_monthly"
425
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_24h"
426
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_6h"
427
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_6h_acc"
428
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.2D_monthly"
429
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.3D_24h"
430
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.3D_6h"
431
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.gr025.3D_monthly"
432
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_24h"
433
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_24h_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_24h_wam"
434
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_6h"
435
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_6h_acc"
436
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_6h_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_6h_wam"
437
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_monthly"
438
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_monthly_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.2D_monthly_wam"
439
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.3D_24h"
440
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.3D_6h"
441
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.atmos.native.3D_monthly"
442
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist-c-lr20-a-0.v20240901.wave.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist-c-lr20-a-0.v20240901.wave.gr025.2D_monthly"
443
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_24h"
444
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_6h"
445
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_6h_acc"
446
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.2D_monthly"
447
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.3D_24h"
448
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.3D_6h"
449
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.gr025.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.gr025.3D_monthly"
450
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_24h"
451
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_24h_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_24h_wam"
452
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_6h"
453
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_6h_acc/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_6h_acc"
454
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_6h_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_6h_wam"
455
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_monthly"
456
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.2D_monthly_wam/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.2D_monthly_wam"
457
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.3D_24h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.3D_24h"
458
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.3D_6h/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.3D_6h"
459
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.atmos.native.3D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.atmos.native.3D_monthly"
460
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco399.hist.v20240901.wave.gr025.2D_monthly/stac"
- type "application/json"
- title "ifs-amip-tco399.hist.v20240901.wave.gr025.2D_monthly"
461
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco3999.hist.v20250101.atmos.gr025.2D_1h/stac"
- type "application/json"
- title "ifs-amip-tco3999.hist.v20250101.atmos.gr025.2D_1h"
462
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco3999.hist.v20250101.atmos.gr025.2D_1h_acc/stac"
- type "application/json"
- title "ifs-amip-tco3999.hist.v20250101.atmos.gr025.2D_1h_acc"
463
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-amip-tco3999.hist.v20250101.atmos.gr025.3D_1h/stac"
- type "application/json"
- title "ifs-amip-tco3999.hist.v20250101.atmos.gr025.3D_1h"
464
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_6hourly_accumulated/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_6hourly_accumulated"
465
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_6hourly_instant/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_6hourly_instant"
466
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_daily_avg"
467
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_daily_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_daily_max"
468
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_daily_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_daily_min"
469
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_monthly_avg"
470
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_monthly_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_monthly_max"
471
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_monthly_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.2D_monthly_min"
472
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.3D_6hourly"
473
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.3D_daily_avg"
474
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.gr025.3D_monthly_avg"
475
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_6hourly_accumulated/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_6hourly_accumulated"
476
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_6hourly_instant/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_6hourly_instant"
477
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_daily_avg"
478
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_daily_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_daily_max"
479
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_daily_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_daily_min"
480
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_monthly_avg"
481
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_monthly_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_monthly_max"
482
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_monthly_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.2D_monthly_min"
483
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.3D_daily_avg"
484
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.atmos.native.3D_monthly_avg"
485
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.land.gr025.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.land.gr025.3D_6hourly"
486
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.land.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.land.gr025.3D_daily_avg"
487
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.land.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.land.gr025.3D_monthly_avg"
488
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.land.native.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.land.native.3D_6hourly"
489
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.land.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.land.native.3D_daily_avg"
490
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.land.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.land.native.3D_monthly_avg"
491
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.2D_daily_avg_1950-2050/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.2D_daily_avg_1950-2050"
492
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.2D_daily_avg_1984-2050/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.2D_daily_avg_1984-2050"
493
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.2D_daily_avg_2000-2050/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.2D_daily_avg_2000-2050"
494
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.AMOC/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.gr025.AMOC"
495
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.native.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.native.2D_daily_avg"
496
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.native.3D_daily_avg"
497
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.eerie-control-1950.v20240304.ocean.native.3D_monthly_avg"
498
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_6hourly_accumulated/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_6hourly_accumulated"
499
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_6hourly_instant/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_6hourly_instant"
500
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_daily_avg"
501
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_daily_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_daily_max"
502
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_daily_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_daily_min"
503
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_monthly_avg"
504
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_monthly_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_monthly_max"
505
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_monthly_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.2D_monthly_min"
506
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.3D_6hourly"
507
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.3D_daily_avg"
508
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.gr025.3D_monthly_avg"
509
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_6hourly_instant/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_6hourly_instant"
510
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_daily_avg"
511
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_daily_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_daily_max"
512
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_daily_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_daily_min"
513
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_monthly_avg"
514
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_monthly_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_monthly_max"
515
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_monthly_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.2D_monthly_min"
516
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.3D_daily_avg"
517
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.atmos.native.3D_monthly_avg"
518
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.land.gr025.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.land.gr025.3D_6hourly"
519
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.land.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.land.gr025.3D_daily_avg"
520
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.land.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.land.gr025.3D_monthly_avg"
521
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.land.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.land.native.3D_daily_avg"
522
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.land.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.land.native.3D_monthly_avg"
523
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.2D_daily_avg"
524
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.2D_monthly_avg"
525
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.3D_daily_avg"
526
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.3D_daily_avg_w/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.3D_daily_avg_w"
527
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.3D_monthly_avg"
528
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.AMOC/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.gr025.AMOC"
529
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.native.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.native.2D_daily_avg"
530
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.native.3D_daily_avg"
531
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.highres-future-ssp245.v20240304.ocean.native.3D_monthly_avg"
532
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_6hourly_accumulated/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_6hourly_accumulated"
533
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_6hourly_instant/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_6hourly_instant"
534
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_daily_avg"
535
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_daily_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_daily_max"
536
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_daily_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_daily_min"
537
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_monthly_avg"
538
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_monthly_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_monthly_max"
539
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_monthly_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.2D_monthly_min"
540
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.3D_6hourly"
541
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.3D_daily_avg"
542
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.gr025.3D_monthly_avg"
543
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_6hourly_accumulated/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_6hourly_accumulated"
544
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_6hourly_instant/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_6hourly_instant"
545
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_6hourly_instant_2/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_6hourly_instant_2"
546
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_daily_avg"
547
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_daily_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_daily_max"
548
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_daily_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_daily_min"
549
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_monthly_avg"
550
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_monthly_max/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_monthly_max"
551
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_monthly_min/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.2D_monthly_min"
552
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.3D_6hourly"
553
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.3D_daily_avg"
554
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.atmos.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.atmos.native.3D_monthly_avg"
555
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.land.gr025.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.land.gr025.3D_6hourly"
556
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.land.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.land.gr025.3D_daily_avg"
557
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.land.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.land.gr025.3D_monthly_avg"
558
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.land.native.3D_6hourly/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.land.native.3D_6hourly"
559
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.land.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.land.native.3D_daily_avg"
560
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.land.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.land.native.3D_monthly_avg"
561
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_daily_avg_1950-2014/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_daily_avg_1950-2014"
562
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_daily_avg_2000-2014/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_daily_avg_2000-2014"
563
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_monthly_avg"
564
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_monthly_avg_mlotst030/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.2D_monthly_avg_mlotst030"
565
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.3D_daily_avg"
566
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.3D_monthly_avg"
567
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.AMOC/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.gr025.AMOC"
568
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.native.2D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.native.2D_daily_avg"
569
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.native.3D_daily_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.native.3D_daily_avg"
570
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-fesom2-sr.hist-1950.v20240304.ocean.native.3D_monthly_avg/stac"
- type "application/json"
- title "ifs-fesom2-sr.hist-1950.v20240304.ocean.native.3D_monthly_avg"
571
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-nemo-er.hist-1950.v20250516.atmos.gr025.Amon/stac"
- type "application/json"
- title "ifs-nemo-er.hist-1950.v20250516.atmos.gr025.Amon"
572
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-nemo-er.hist-1950.v20250516.atmos.gr025.Omon/stac"
- type "application/json"
- title "ifs-nemo-er.hist-1950.v20250516.atmos.gr025.Omon"
573
- rel "child"
- href "https://eerie.cloud.dkrz.de/datasets/ifs-nemo-er.hist-1950.v20250516.atmos.gr025.SImon/stac"
- type "application/json"
- title "ifs-nemo-er.hist-1950.v20250516.atmos.gr025.SImon"
- title "ESM data from DKRZ in Zarr format"
extent
spatial
bbox[] 1 items
0[] 4 items
- 0 -180
- 1 -90
- 2 180
- 3 90
temporal
interval[] 1 items
0[] 2 items
- 0 "1850-01-01T00:00:00Z"
- 1 "2101-01-01T00:00:00Z"
- license "other"
keywords[] 9 items
- 0 "EERIE"
- 1 "cloud"
- 2 "ICON"
- 3 "NextGEMs"
- 4 "ERA5"
- 5 "IFS"
- 6 "FESOM"
- 7 "NEMO"
- 8 "HadGEM"
providers[] 1 items
0
- name "DKRZ"
- description "The data host of eerie.cloud"
roles[] 1 items
- 0 "host"
- url "https://dkrz.de"
assets
doc
- href "https://pad.gwdg.de/OZo5HMC4R6iljvZHlo-BzQ#"
- type "text/html"
- title "Technical documentation"
roles[] 1 items
- 0 "OVERVIEW"
Each child link in the collection corresponds to a dataset and is formatted as a STAC item. Datasets of the km-scale-cloud are highly aggregated.
We can do:
dataset_items=[
pystac.Item.from_file(link.href)
for link in kmscale_collection.get_links("child")
]
item=dataset_items[0]
item
- type "Feature"
- stac_version "1.1.0"
stac_extensions[] 3 items
- 0 "https://stac-extensions.github.io/xarray-assets/v1.0.0/schema.json"
- 1 "https://stac-extensions.github.io/datacube/v2.2.0/schema.json"
- 2 "https://stac-extensions.github.io/alternate-assets/v1.2.0/schema.json"
- id "cosmo-rea-1hrPt_atmos"
geometry
- type "Polygon"
coordinates[] 1 items
0[] 5 items
0[] 2 items
- 0 -180.0
- 1 -90.0
1[] 2 items
- 0 -180.0
- 1 90.0
2[] 2 items
- 0 180.0
- 1 90.0
3[] 2 items
- 0 180.0
- 1 -90.0
4[] 2 items
- 0 -180.0
- 1 -90.0
bbox[] 4 items
- 0 -180.0
- 1 -90.0
- 2 180.0
- 3 90.0
properties
- title "cosmo-rea-1hrPt_atmos"
- description " Simulation data from project 'COSMO-REA' produced by Earth System Model 'COSMO' and run by institution 'DWD' for the experiment 'REA6' You can use this dataset in xarray. ``` import xarray as xr import requests item=requests.get("https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/stac").json() asset="dkrz-disk" #asset="eerie-cloud" # from everywhere else xr.open_dataset( item["assets"][asset]["href"], **item["assets"][asset]["xarray:open_kwargs"], storage_options=item["assets"][asset].get("xarray:storage_options") ) ``` # COSMO-REA6 regional reanalysis data prepared for ESGF. Simulation data from project 'COSMO-REA' produced by Earth System Model 'COSMO' and run by institution 'DWD' for the experiment 'REA6'"
- created "2026-02-19T00:00:21Z"
keywords[] 2 items
- 0 "COSMO"
- 1 "REA6 regional reanalysis data prepared for ESGF."
providers[] 2 items
0
- name "DKRZ"
- description "The data host of the items and assets"
roles[] 1 items
- 0 "host"
- url "https://dkrz.de"
1
- name "DWD"
- description "Deutscher Wetterdienst, Offenbach am Main 63067, Germany"
roles[] 1 items
- 0 "producer"
- url "N/A"
- license "other"
variables[] 124 items
- 0 "ad100m"
- 1 "ad125m"
- 2 "ad150m"
- 3 "ad175m"
- 4 "ad200m"
- 5 "ad40m"
- 6 "ad60m"
- 7 "ad80m"
- 8 "clh"
- 9 "clivi"
- 10 "cll"
- 11 "clm"
- 12 "clt"
- 13 "clwvi"
- 14 "hfls"
- 15 "hfss"
- 16 "hurs"
- 17 "hus"
- 18 "hus100"
- 19 "hus1000"
- 20 "hus150"
- 21 "hus200"
- 22 "hus250"
- 23 "hus300"
- 24 "hus400"
- 25 "hus50"
- 26 "hus500"
- 27 "hus600"
- 28 "hus70"
- 29 "hus700"
- 30 "hus750"
- 31 "hus850"
- 32 "hus925"
- 33 "huss"
- 34 "orog"
- 35 "pfull"
- 36 "prw"
- 37 "ps"
- 38 "psl"
- 39 "rlds"
- 40 "rlus"
- 41 "rlut"
- 42 "rsds"
- 43 "rsdsdir"
- 44 "rsdt"
- 45 "rsus"
- 46 "rsut"
- 47 "sfcWind"
- 48 "ta"
- 49 "ta100"
- 50 "ta1000"
- 51 "ta150"
- 52 "ta200"
- 53 "ta250"
- 54 "ta300"
- 55 "ta400"
- 56 "ta50"
- 57 "ta500"
- 58 "ta600"
- 59 "ta70"
- 60 "ta700"
- 61 "ta750"
- 62 "ta850"
- 63 "ta925"
- 64 "tas"
- 65 "tauu"
- 66 "tauv"
- 67 "tdps"
- 68 "tke"
- 69 "ts"
- 70 "twp"
- 71 "ua"
- 72 "ua100"
- 73 "ua1000"
- 74 "ua150"
- 75 "ua200"
- 76 "ua250"
- 77 "ua300"
- 78 "ua400"
- 79 "ua50"
- 80 "ua500"
- 81 "ua600"
- 82 "ua70"
- 83 "ua700"
- 84 "ua750"
- 85 "ua850"
- 86 "ua925"
- 87 "uas"
- 88 "va"
- 89 "va100"
- 90 "va1000"
- 91 "va150"
- 92 "va200"
- 93 "va250"
- 94 "va300"
- 95 "va400"
- 96 "va50"
- 97 "va500"
- 98 "va600"
- 99 "va70"
- 100 "va700"
- 101 "va750"
- 102 "va850"
- 103 "va925"
- 104 "vas"
- 105 "wd100m"
- 106 "wd10m"
- 107 "wd125m"
- 108 "wd150m"
- 109 "wd175m"
- 110 "wd200m"
- 111 "wd40m"
- 112 "wd60m"
- 113 "wd80m"
- 114 "ws100m"
- 115 "ws125m"
- 116 "ws150m"
- 117 "ws175m"
- 118 "ws200m"
- 119 "ws40m"
- 120 "ws60m"
- 121 "ws80m"
- 122 "zf"
- 123 "zmla"
cube:dimensions
time
- type "temporal"
extent[] 2 items
- 0 "1995-01-01T01:00:00Z"
- 1 "2019-09-01T00:00:00Z"
cube:variables
ad100m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 100m"
ad125m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 125m"
ad150m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 150m"
ad175m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 175m"
ad200m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 200m"
ad40m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 40m"
ad60m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 60m"
ad80m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-3"
- description "Air Density at 80m"
clh
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "%"
- description "High Level Cloud Area Percentage"
clivi
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-2"
- description "Ice Water Path"
cll
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "%"
- description "Low Level Cloud Cover Percentage"
clm
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "%"
- description "Mid Level Cloud Cover Percentage"
clt
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "%"
- description "Total Cloud Cover Percentage"
clwvi
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-2"
- description "Condensed Water Path"
hfls
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Upward Latent Heat Flux"
hfss
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Upward Sensible Heat Flux"
hurs
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "%"
- description "Near-Surface Relative Humidity"
hus
- type "data"
dimensions[] 4 items
- 0 "time"
- 1 "lev"
- 2 "rlat"
- 3 "rlon"
- unit "1"
- description "Specific Humidity"
hus100
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus1000
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus150
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus200
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus250
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus300
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus400
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus50
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus500
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus600
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus70
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus700
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus750
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus850
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
hus925
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Specific Humidity"
huss
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "1"
- description "Near-Surface Specific Humidity"
orog
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m"
- description "Surface Altitude"
pfull
- type "data"
dimensions[] 4 items
- 0 "time"
- 1 "lev"
- 2 "rlat"
- 3 "rlon"
- unit "Pa"
- description "Pressure at Model Full-Levels"
prw
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-2"
- description "Water Vapor Path"
ps
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "Pa"
- description "Surface Air Pressure"
psl
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "Pa"
- description "Sea Level Pressure"
rlds
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Downwelling Longwave Radiation"
rlus
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Upwelling Longwave Radiation"
rlut
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "TOA Outgoing Longwave Radiation"
rsds
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Downwelling Shortwave Radiation"
rsdsdir
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Direct Downwelling Shortwave Radiation"
rsdt
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "TOA Incident Shortwave Radiation"
rsus
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "Surface Upwelling Shortwave Radiation"
rsut
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "W m-2"
- description "TOA Outgoing Shortwave Radiation"
sfcWind
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Hourly Near-Surface Wind Speed"
ta
- type "data"
dimensions[] 4 items
- 0 "time"
- 1 "lev"
- 2 "rlat"
- 3 "rlon"
- unit "K"
- description "Air Temperature"
ta100
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta1000
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta150
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta200
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta250
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta300
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta400
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta50
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta500
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta600
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta70
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta700
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta750
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta850
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
ta925
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Air Temperature"
tas
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Near-Surface Air Temperature"
tauu
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "Pa"
- description "Surface Downward Eastward Wind Stress"
tauv
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "Pa"
- description "Surface Downward Northward Wind Stress"
tdps
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "2m Dewpoint Temperature"
tke
- type "data"
dimensions[] 4 items
- 0 "time"
- 1 "lev"
- 2 "rlat"
- 3 "rlon"
- unit "m2 s-2"
- description "Turbulent Kinetic Energy"
ts
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "K"
- description "Surface Temperature"
twp
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "kg m-2"
- description "Total Water Path"
ua
- type "data"
dimensions[] 4 items
- 0 "time"
- 1 "lev"
- 2 "rlat"
- 3 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua100
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua1000
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua150
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua200
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua250
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua300
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua400
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua50
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua500
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua600
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua70
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua700
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua750
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua850
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
ua925
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Wind"
uas
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Eastward Near-Surface Wind"
va
- type "data"
dimensions[] 4 items
- 0 "time"
- 1 "lev"
- 2 "rlat"
- 3 "rlon"
- unit "m s-1"
- description "Northward Wind"
va100
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va1000
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va150
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va200
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va250
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va300
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va400
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va50
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va500
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va600
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va70
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va700
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va750
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va850
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
va925
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Wind"
vas
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Northward Near-Surface Wind"
wd100m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 100m"
wd10m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 10m"
wd125m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 125m"
wd150m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 150m"
wd175m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 175m"
wd200m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 200m"
wd40m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 40m"
wd60m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 60m"
wd80m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "degree"
- description "Wind From Direction at 80m"
ws100m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 100m"
ws125m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 125m"
ws150m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 150m"
ws175m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 175m"
ws200m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 200m"
ws40m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 40m"
ws60m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 60m"
ws80m
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m s-1"
- description "Wind Speed at 80m"
zf
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m"
- description "Freezing Level Altitude"
zmla
- type "data"
dimensions[] 3 items
- 0 "time"
- 1 "rlat"
- 2 "rlon"
- unit "m"
- description "Height of Boundary Layer"
- CDO "Climate Data Operators version 2.2.1 (https://mpimet.mpg.de/cdo)"
- CORDEX_domain "EUR-6km"
- Conventions "CF-1.7"
- activity_id "COSMO-REA"
- cmor_version "3.6.0"
- contact "cdc.rea@dwd.de"
- creation_date "2026-02-19T00:00:21Z"
- data_specs_version "01.01.00"
- driving_experiment "ECMWF-ERAINT, global reanalysis"
- driving_experiment_name "global reanalysis"
- driving_model_ensemble_member "r1i1p1f1"
- driving_model_id "ECMWF-ERAINT"
- experiment "regional reanalysis"
- experiment_id "REA6"
- external_variables "areacella"
- forcing_index 1
- frequency "1hr"
- grid "gn"
- grid_label "gn"
- initialization_index 1
- institution "Deutscher Wetterdienst, Offenbach am Main 63067, Germany"
- institution_id "DWD"
- mip_era "CMIP6"
- model_id "COSMO"
- physics_index 1
- product "reanalysis"
- project_id "COSMO-REA"
- rcm_version_id "v1"
- realization_index 1
- realm "atmos"
- references "Reanalysis conducted in cooperation with Hans-Ertel-Centre. Bollmeyer, C., Keller, J. D., Ohlwein, C., Wahl, S., Crewell, S., Friederichs, P., Hense, A., Keune, J., Kneifel, S., Pscheidt, I., Redl, S., and Steinke, S.: Towards a high-resolution regional reanalysis for the European CORDEX domain, Q. J. R. Meteorol. Soc., 141, 1–15, 2015, https://doi.org/10.1002/qj.2486. Kaspar, F., Niermann, D., Borsche, M., Fiedler, S., Keller, J., Potthast, R., Rösch, T., Spangehl, T., and Tinz, B.: Regional atmospheric reanalysis activities at Deutscher Wetterdienst: review of evaluation results and application examples with a focus on renewable energy, Adv. Sci. Res., 17, 115–128, https://doi.org/10.5194/asr-17-115-2020, 2020."
- source "COSMO v4.25 (2012): aerosol: none/prescribed atmos: COnsortium for Small-Scale MOdelling (COSMO) limited-area model (LAM) version 4.25 (Arakawa-C/Lorenz Grid; 848 x 824 longitude/latitude; 40 levels; top level 40 hPa) atmosChem: none land: TERRA-ML landIce: none/prescribed ocean: none/prescribed ocnBgchem: none seaIce: none/prescribed solidLand: none/prescribed"
- source_id "COSMO"
- source_type "AGCM"
- sub_experiment "none"
- sub_experiment_id "none"
- table_id "1hrPt"
- table_info "Creation Date:(01 Sep 2023) MD5:da644e15786f7b8daa83bbad28a0d9be"
- tracking_id "hdl:21.14103/428f45ee-5c29-4d09-97a5-351fb45c507a"
- variable_id "tauu"
- variant_info "Regional reanalysis forced by ERA-Interim."
- variant_label "r1i1p1f1"
- _xpublish_id "cosmo-rea-1hrPt_atmos"
- start_datetime "1995-01-01T01:00:00Z"
- end_datetime "2019-09-01T00:00:00Z"
- datetime None
asset_types[] 2 items
- 0 "dkrz-disk"
- 1 "eerie-cloud"
links[] 2 items
0
- rel "self"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/stac"
- type "application/json"
1
- rel "collection"
- href "https://eerie.cloud.dkrz.de/stac-collection-all.json"
- type "application/json"
assets
dkrz-disk
- href "reference::file:///work/bm1344/DKRZ/git/cosmo-rea-kerchunks/kerchunks/file/parquets/1hrPt_atmos/combined.parq"
- type "application/vnd+zarr"
- title "Zarr-access from Lustre"
- description "Chunk-based access on Raw Data"
xarray:storage_options
xarray:storage_options
- lazy True
xarray:open_kwargs
- engine "zarr"
- chunks "auto"
- consolidated False
roles[] 1 items
- 0 "data"
eerie-cloud
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/kerchunk"
- type "application/vnd+zarr"
- title "Zarr-access through Eerie Cloud"
- description "Chunk-based access on raw-encoded data"
- Volume "86675 GB uncompressed"
- No of data variables "124"
xarray:open_kwargs
- engine "zarr"
- chunks "auto"
- consolidated True
- xarray:storage_options None
alternate
processed
- name "Rechunked and uniformly compressed data"
- description "Server-side on-the-fly rechunked and unifromly encoded data"
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/zarr"
roles[] 1 items
- 0 "data"
xarray_view
- href "https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/"
- type "text/html"
- title "Xarray dataset"
- description "HTML representation of the xarray dataset"
roles[] 1 items
- 0 "overview"
jupyterlite
- href "https://swift.dkrz.de/v1/dkrz_7fa6baba-db43-4d12-a295-8e3ebb1a01ed/apps/jupyterlite/index.html"
- type "text/html"
- title "Jupyterlite access"
- description "Web-assembly based analysis platform with access to this item"
roles[] 1 items
- 0 "analysis"
gridlook
- href "https://gridlook.pages.dev/#https://eerie.cloud.dkrz.de/datasets/cosmo-rea-1hrPt_atmos/stac"
- type "text/html"
- title "Visualization with gridlook"
- description "Visualization with gridlook"
roles[] 1 items
- 0 "Visualization"
providers[] 1 items
0
- name "DKRZ"
- description "The data host of the items and assets"
roles[] 1 items
- 0 "host"
- url "https://dkrz.de"
- default_var "tas"
- name "COSMO-REA6 regional reanalysis data prepared for ESGF."
levels[] 1 items
0
- name "cosmo-rea-1hrPt_atmos"
time
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/zarr"
grid
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
datasources
ad100m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad125m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad150m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad175m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad200m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad40m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad60m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ad80m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
clh
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
clivi
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
cll
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
clm
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
clt
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
clwvi
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hfls
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hfss
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hurs
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus100
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus1000
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus150
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus200
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus250
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus300
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus400
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus50
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus500
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus600
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus70
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus700
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus750
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus850
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
hus925
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
huss
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
orog
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
pfull
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
prw
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ps
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
psl
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rlds
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rlus
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rlut
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rsds
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rsdsdir
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rsdt
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rsus
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rsut
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
sfcWind
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta100
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta1000
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta150
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta200
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta250
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta300
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta400
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta50
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta500
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta600
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta70
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta700
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta750
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta850
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ta925
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
tas
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
tauu
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
tauv
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
tdps
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
tke
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ts
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
twp
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua100
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua1000
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua150
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua200
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua250
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua300
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua400
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua50
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua500
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua600
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua70
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua700
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua750
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua850
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ua925
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
uas
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va100
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va1000
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va150
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va200
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va250
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va300
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va400
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va50
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va500
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va600
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va70
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va700
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va750
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va850
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
va925
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
vas
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd100m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd10m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd125m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd150m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd175m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd200m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd40m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd60m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
wd80m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws100m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws125m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws150m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws175m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws200m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws40m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws60m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
ws80m
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
zf
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
zmla
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
height
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
latitude
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
lev
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
longitude
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rlat
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rlon
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
rotated_latitude_longitude
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
time
- store "https://eerie.cloud.dkrz.de/datasets/"
- dataset "cosmo-rea-1hrPt_atmos/kerchunk"
We can use properties or ids to browse and search the items:
selection=[
a
for a in dataset_items
if all(b in a.id.lower() for b in ["icon","gr025","monthly","2d","hist","atmos"])
]
item=selection[0]
len(selection)
1
Each dataset item has multiple assets corresponding to different ways how to open the dataset, e.g. with “dkrz-disk” should be used when working on levante.
We now use the “eerie-cloud” asset. It provides information on volume and number of variables for the uncompressed, total dataset.
asset="eerie-cloud"
ec_asset=item.get_assets()[asset]
ec_asset_dict=ec_asset.to_dict()
ec_asset
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_monthly_mean/kerchunk"
- type "application/vnd+zarr"
- title "Zarr-access through Eerie Cloud"
- description "Chunk-based access on raw-encoded data"
- Volume "100 GB uncompressed"
- No of data variables "33"
xarray:open_kwargs
- engine "zarr"
- chunks "auto"
- consolidated True
- xarray:storage_options None
alternate
processed
- name "Rechunked and uniformly compressed data"
- description "Server-side on-the-fly rechunked and unifromly encoded data"
- href "https://eerie.cloud.dkrz.de/datasets/icon-esm-er.hist-1950.v20240618.atmos.gr025.2d_monthly_mean/zarr"
roles[] 1 items
- 0 "data"
We can open datasets with the asset:
import xarray as xr
zarr_uri=ec_asset_dict["href"]
xr.open_dataset(
zarr_uri,
**ec_asset_dict["xarray:open_kwargs"],
storage_options=ec_asset_dict.get("xarray:storage_options")
)
<xarray.Dataset> Size: 107GB
Dimensions: (time: 780, lat: 721, lon: 1440, height_3: 1, bnds: 2,
height: 1, height_2: 1)
Coordinates:
* time (time) datetime64[ns] 6kB 1950-01-31T23:59:59 ... 2014-12-...
* lat (lat) float64 6kB -90.0 -89.75 -89.5 ... 89.5 89.75 90.0
* lon (lon) float64 12kB 0.0 0.25 0.5 0.75 ... 359.2 359.5 359.8
* height_3 (height_3) float64 8B 90.0
* height (height) float64 8B 2.0
* height_2 (height_2) float64 8B 10.0
Dimensions without coordinates: bnds
Data variables: (12/34)
clivi (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
cllvi (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
clt (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
evspsbl (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
height_3_bnds (time, height_3, bnds) float64 12kB dask.array<chunksize=(780, 1, 2), meta=np.ndarray>
hfls (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
... ...
tasmin (time, height, lat, lon) float32 3GB dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
tauu (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
tauv (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
ts (time, lat, lon) float32 3GB dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
uas (time, height_2, lat, lon) float32 3GB dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
vas (time, height_2, lat, lon) float32 3GB dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
Attributes: (12/31)
Conventions: CF-1.7 CMIP-6.2
activity_id: HighResMIP
data_specs_version: 01.00.32
forcing_index: 1
initialization_index: 1
license: EERIE model data produced by MPI-M is licensed und...
... ...
parent_activity_id: HighResMIP
sub_experiment_id: none
experiment: coupled historical 1950-2014
source: ICON-ESM-ER (2023): \naerosol: none, prescribed MA...
institution: Max Planck Institute for Meteorology, Hamburg 2014...
sub_experiment: none- time: 780
- lat: 721
- lon: 1440
- height_3: 1
- bnds: 2
- height: 1
- height_2: 1
- time(time)datetime64[ns]1950-01-31T23:59:59 ... 2014-12-...
- standard_name :
- time
array(['1950-01-31T23:59:59.000000000', '1950-02-28T23:59:59.000000000', '1950-03-31T23:59:59.000000000', ..., '2014-10-31T23:59:59.000000000', '2014-11-30T23:59:59.000000000', '2014-12-31T23:59:59.000000000'], shape=(780,), dtype='datetime64[ns]') - lat(lat)float64-90.0 -89.75 -89.5 ... 89.75 90.0
- axis :
- Y
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
array([-90. , -89.75, -89.5 , ..., 89.5 , 89.75, 90. ], shape=(721,))
- lon(lon)float640.0 0.25 0.5 ... 359.2 359.5 359.8
- axis :
- X
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([0.0000e+00, 2.5000e-01, 5.0000e-01, ..., 3.5925e+02, 3.5950e+02, 3.5975e+02], shape=(1440,)) - height_3(height_3)float6490.0
- axis :
- Z
- bounds :
- height_3_bnds
- long_name :
- generalized_height
- standard_name :
- height
array([90.])
- height(height)float642.0
- axis :
- Z
- long_name :
- height
- positive :
- up
- standard_name :
- height
- units :
- m
array([2.])
- height_2(height_2)float6410.0
- axis :
- Z
- long_name :
- height
- positive :
- up
- standard_name :
- height
- units :
- m
array([10.])
- clivi(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- cloud ice path
- param :
- 70.1.0
- standard_name :
- atmosphere_mass_content_of_cloud_ice
- units :
- kg m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - cllvi(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- cloud liquid water path
- param :
- 69.1.0
- standard_name :
- atmosphere_mass_content_of_cloud_liquid_water
- units :
- kg m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - clt(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- total cloud cover
- param :
- 1.6.0
- standard_name :
- clt
- units :
- m2 m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - evspsbl(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- evaporation
- param :
- 6.1.0
- standard_name :
- evap
- units :
- kg m-2 s-1
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - height_3_bnds(time, height_3, bnds)float64dask.array<chunksize=(780, 1, 2), meta=np.ndarray>
Array Chunk Bytes 12.19 kiB 12.19 kiB Shape (780, 1, 2) (780, 1, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hfls(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- latent heat flux
- param :
- 10.0.0
- standard_name :
- lhflx
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - hfss(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- sensible heat flux
- param :
- 11.0.0
- standard_name :
- shflx
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - hur(time, height_3, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- relative humidity
- param :
- 1.1.0
- standard_name :
- relative_humidity
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - hus2m(time, height, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- specific humidity in 2m
- param :
- 6.0.0
- standard_name :
- qv2m
- units :
- kg kg-1
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - pr(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- precipitation flux
- param :
- 52.1.0
- standard_name :
- pr
- units :
- kg m-2 s-1
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - prw(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- water vapor path
- param :
- 64.1.0
- standard_name :
- atmosphere_mass_content_of_water_vapor
- units :
- kg m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - ps(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface pressure
- param :
- 0.3.0
- standard_name :
- surface_air_pressure
- units :
- Pa
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - psl(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- mean sea level pressure
- param :
- 1.3.0
- standard_name :
- mean sea level pressure
- units :
- Pa
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rlds(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface downwelling longwave radiation
- param :
- 3.5.0
- standard_name :
- surface_downwelling_longwave_flux_in_air
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rldscs(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface downwelling clear-sky longwave radiation
- param :
- 203.5.0
- standard_name :
- surface_downwelling_longwave_flux_in_air_assuming_clear_sky
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rlus(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface upwelling longwave radiation
- param :
- 199.5.0
- standard_name :
- surface_upwelling_longwave_flux_in_air
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rlut(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- level_type :
- toa
- long_name :
- toa outgoing longwave radiation
- param :
- 4.5.0
- standard_name :
- toa_outgoing_longwave_flux
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rlutcs(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- level_type :
- toa
- long_name :
- toa outgoing clear-sky longwave radiation
- param :
- 204.5.0
- standard_name :
- toa_outgoing_longwave_flux_assuming_clear_sky
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsds(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface downwelling shortwave radiation
- param :
- 7.4.0
- standard_name :
- surface_downwelling_shortwave_flux_in_air
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsdscs(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface downwelling clear-sky shortwave radiation
- param :
- 207.4.0
- standard_name :
- surface_downwelling_shortwave_flux_in_air_assuming_clear_sky
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsdt(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- level_type :
- toa
- long_name :
- toa incident shortwave radiation
- param :
- 201.4.0
- standard_name :
- toa_incoming_shortwave_flux
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsus(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface upwelling shortwave radiation
- param :
- 199.4.0
- standard_name :
- surface_upwelling_shortwave_flux_in_air
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsuscs(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface upwelling clear-sky shortwave radiation
- param :
- 209.4.0
- standard_name :
- surface_upwelling_shortwave_flux_in_air_assuming_clear_sky
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsut(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- level_type :
- toa
- long_name :
- toa outgoing shortwave radiation
- param :
- 8.4.0
- standard_name :
- toa_outgoing_shortwave_flux
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - rsutcs(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- level_type :
- toa
- long_name :
- toa outgoing clear-sky shortwave radiation
- param :
- 208.4.0
- standard_name :
- toa_outgoing_shortwave_flux_assuming_clear_sky
- units :
- W m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - sfcwind(time, height_2, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- 10m windspeed
- param :
- 1.2.0
- standard_name :
- sfcwind
- units :
- m s-1
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - tas(time, height, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- temperature in 2m
- param :
- 0.0.0
- standard_name :
- tas
- units :
- K
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - tasmax(time, height, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- maximum 2m temperature
- param :
- 4.0.0
- standard_name :
- tasmax
- units :
- K
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - tasmin(time, height, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- minimum 2m temperature
- param :
- 5.0.0
- standard_name :
- tasmin
- units :
- K
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - tauu(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- u-momentum flux at the surface
- param :
- 17.2.0
- standard_name :
- u_stress
- units :
- N m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - tauv(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- v-momentum flux at the surface
- param :
- 18.2.0
- standard_name :
- v_stress
- units :
- N m-2
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - ts(time, lat, lon)float32dask.array<chunksize=(32, 721, 1440), meta=np.ndarray>
- long_name :
- surface temperature
- param :
- 0.0.0
- standard_name :
- surface_temperature
- units :
- K
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 721, 1440) (32, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - uas(time, height_2, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- zonal wind in 10m
- param :
- 2.2.0
- standard_name :
- uas
- units :
- m s-1
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray - vas(time, height_2, lat, lon)float32dask.array<chunksize=(32, 1, 721, 1440), meta=np.ndarray>
- long_name :
- meridional wind in 10m
- param :
- 3.2.0
- standard_name :
- vas
- units :
- m s-1
Array Chunk Bytes 3.02 GiB 126.74 MiB Shape (780, 1, 721, 1440) (32, 1, 721, 1440) Dask graph 25 chunks in 2 graph layers Data type float32 numpy.ndarray
- Conventions :
- CF-1.7 CMIP-6.2
- activity_id :
- HighResMIP
- data_specs_version :
- 01.00.32
- forcing_index :
- 1
- initialization_index :
- 1
- license :
- EERIE model data produced by MPI-M is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses). The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- EERIE
- project_id :
- EERIE
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- source_type :
- AOGCM
- variant_label :
- r1i1p1f1
- creation_date :
- 2025-04-02T13:48:28
- institution_id :
- MPI-M
- source_id :
- ICON-ESM-ER
- experiment_id :
- hist-1950
- contact :
- juergen.kroeger@mpimet.mpg.de
- references :
- Hohenegger et al., ICON-Sapphire: simulating the components of the Earth system and their interactions at kilometer and subkilometer scales. Geosci. Model Dev., 16, 779–811, 2023, https://doi.org/10.5194/gmd-16-779-2023
- nominal_resolution :
- 10 km
- version_id :
- v20240618
- realm :
- atmos
- grid :
- gr025
- grid_label :
- gr
- parent_experiment_id :
- spinup-1950
- parent_activity_id :
- HighResMIP
- sub_experiment_id :
- none
- experiment :
- coupled historical 1950-2014
- source :
- ICON-ESM-ER (2023): aerosol: none, prescribed MACv2-SP atmos: ICON-A (icosahedral/triangles; 10 km; 90 levels; top level 80 km) atmosChem: none land: JSBACH4.20 landIce: none/prescribed ocean: ICON-O (icosahedral/triangles; 5 km; 72 levels; top grid cell 0-2 m) ocnBgchem: none seaIce: unnamed (thermodynamic (Semtner zero-layer) dynamic (Hibler 79) sea ice model)
- institution :
- Max Planck Institute for Meteorology, Hamburg 20146, Germany
- sub_experiment :
- none
Advanced: Retrievel setting#
Before we start excessively transfering data chunks, we need to understand how the km-scale cloud manages to prevent us from running into rejected requests. The km-scale-cloud aims to serve data to multiple users at once. Therefore, it allows 200 requests per client IP before it burst new requests and returns a 429 error. It also only allows 60 requests per datasets.
On the other side, the python Zarr that we use for access is embarassingly parallel. Under the hood, it uses the aiohttp client which limits the number of simultanoues request to 100 per session.
Thus, with default settings, you can only retrieve data from 2 datasets (2 * 100) at once! Moreover, it make sense to configure the client so that it uses less requests at once. A reasonable number of concurrent requests depends on
your network bandwidth
the chunk size of the target data For some high resolution data (GRIB formatted), one chunk can be large (>>10MB). In that case, you already reach a performance that fill your network capacity with only a few parallel chunks.
This is how parallel requests are limited to 2:
import aiohttp
so_limited={
"client_kwargs": {
"connector": aiohttp.TCPConnector(limit=2),
}
}
xr.open_dataset(
zarr_uri,
**ec_asset_dict["xarray:open_kwargs"],
storage_options=so_limited
)
Advanced: Two Zarr versions#
The km-scale-cloud provides two Zar access endpoints for the same datasets which differ:
/kerchunk suffix: The preferred href
delivers data as stored in the original without any processing on the server. Original zarr data, e.g. NextGEMs ICON, is just passed through to the user.
/zarr suffix: The alternate href
delivers data processed on server-side with dask. The resulting data is
rechunked in large, dask-optimal chunksizes (O(100MB) in memory)
uniformly compressed with blosc. Native data is lossy compressed with 12-bit bitrouding.
Kerchunk have a smaller footprint in both memory and compute on the server. zarr reduces data volume before transfer to support users with limited network bandwidth.
Intake#
Warning
The Intake catalog is a legacy interface and requires intake<2
The server dynamically creates an intake catalog which is synced with the available dataset.
import intake
cat = intake.open_catalog("https://eerie.cloud.dkrz.de/intake.yaml")
We can list all datasets available in the catalog with list:
all_dkrz_datasets = list(cat)
len(all_dkrz_datasets)
571
The two zarr access points for each dataset are given in the description:
dsname = all_dkrz_datasets[-1]
cat[dsname].describe()
{'name': 'tape_test',
'container': 'xarray',
'plugin': ['zarr'],
'driver': ['zarr'],
'description': '',
'direct_access': 'forbid',
'user_parameters': [{'name': 'method',
'description': 'server-side loading method',
'type': 'str',
'allowed': ['kerchunk', 'zarr'],
'default': 'kerchunk'}],
'metadata': {},
'args': {'consolidated': True,
'urlpath': 'https://eerie.cloud.dkrz.de/datasets/tape_test/{{ method }}'}}
to_dask() opens the dataset:
cat[dsname].to_dask()
<xarray.Dataset> Size: 12TB
Dimensions: (time: 569784, ncells: 5242880)
Coordinates:
* time (time) datetime64[ns] 5MB 1950-01-01T00:59:59 ... 201...
cell_sea_land_mask (ncells) int32 21MB dask.array<chunksize=(5242880,), meta=np.ndarray>
lat (ncells) float64 42MB dask.array<chunksize=(5242880,), meta=np.ndarray>
lon (ncells) float64 42MB dask.array<chunksize=(5242880,), meta=np.ndarray>
Dimensions without coordinates: ncells
Data variables:
rlut (time, ncells) float32 12TB dask.array<chunksize=(1, 5000000), meta=np.ndarray>
Attributes: (12/31)
Conventions: CF-1.7 CMIP-6.2
activity_id: HighResMIP
contact: juergen.kroeger@mpimet.mpg.de
creation_date: 2025-04-02T13:48:28
data_specs_version: 01.00.32
experiment: coupled historical 1950-2014
... ...
source_id: ICON-ESM-ER
source_type: AOGCM
sub_experiment: none
sub_experiment_id: none
variant_label: r1i1p1f1
version_id: v20240618- time: 569784
- ncells: 5242880
- time(time)datetime64[ns]1950-01-01T00:59:59 ... 2014-12-...
array(['1950-01-01T00:59:59.000000000', '1950-01-01T01:59:59.000000000', '1950-01-01T02:59:59.000000000', ..., '2014-12-31T21:59:59.000000000', '2014-12-31T22:59:59.000000000', '2014-12-31T23:59:59.000000000'], shape=(569784,), dtype='datetime64[ns]') - cell_sea_land_mask(ncells)int32dask.array<chunksize=(5242880,), meta=np.ndarray>
- long_name :
- sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell
- units :
- 2,1,-1,-
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
- dtype :
- int32
- zlib :
- False
- szip :
- False
- zstd :
- False
- bzip2 :
- False
- blosc :
- False
- shuffle :
- False
- complevel :
- 0
- fletcher32 :
- False
- contiguous :
- True
- chunksizes :
- None
- source :
- /work/bm1344/DKRZ/kerchunks_batched/erc2002/icon_grid_0033_R02B08_G.nc
- original_shape :
- [5242880]
Array Chunk Bytes 20.00 MiB 20.00 MiB Shape (5242880,) (5242880,) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - lat(ncells)float64dask.array<chunksize=(5242880,), meta=np.ndarray>
- long_name :
- center latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- clat_vertices
- dtype :
- float64
- zlib :
- False
- szip :
- False
- zstd :
- False
- bzip2 :
- False
- blosc :
- False
- shuffle :
- False
- complevel :
- 0
- fletcher32 :
- False
- contiguous :
- True
- chunksizes :
- None
- source :
- /work/bm1344/DKRZ/kerchunks_batched/erc2002/icon_grid_0033_R02B08_G.nc
- original_shape :
- [5242880]
Array Chunk Bytes 40.00 MiB 40.00 MiB Shape (5242880,) (5242880,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lon(ncells)float64dask.array<chunksize=(5242880,), meta=np.ndarray>
- long_name :
- center longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- clon_vertices
- dtype :
- float64
- zlib :
- False
- szip :
- False
- zstd :
- False
- bzip2 :
- False
- blosc :
- False
- shuffle :
- False
- complevel :
- 0
- fletcher32 :
- False
- contiguous :
- True
- chunksizes :
- None
- source :
- /work/bm1344/DKRZ/kerchunks_batched/erc2002/icon_grid_0033_R02B08_G.nc
- original_shape :
- [5242880]
Array Chunk Bytes 40.00 MiB 40.00 MiB Shape (5242880,) (5242880,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray
- rlut(time, ncells)float32dask.array<chunksize=(1, 5000000), meta=np.ndarray>
- CDI_grid_type :
- unstructured
- level_type :
- toa
- long_name :
- toa outgoing longwave radiation
- number_of_grid_in_reference :
- 1
- param :
- 4.5.0
- standard_name :
- toa_outgoing_longwave_flux
- units :
- W m-2
Array Chunk Bytes 10.87 TiB 19.07 MiB Shape (569784, 5242880) (1, 5000000) Dask graph 1139568 chunks in 2 graph layers Data type float32 numpy.ndarray
- Conventions :
- CF-1.7 CMIP-6.2
- activity_id :
- HighResMIP
- contact :
- juergen.kroeger@mpimet.mpg.de
- creation_date :
- 2025-04-02T13:48:28
- data_specs_version :
- 01.00.32
- experiment :
- coupled historical 1950-2014
- experiment_id :
- hist-1950
- forcing_index :
- 1
- grid :
- native
- grid_label :
- gn
- initialization_index :
- 1
- institution :
- Max Planck Institute for Meteorology, Hamburg 20146, Germany
- institution_id :
- MPI-M
- license :
- EERIE model data produced by MPI-M is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses). The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- EERIE
- nominal_resolution :
- 10 km
- parent_activity_id :
- HighResMIP
- parent_experiment_id :
- spinup-1950
- physics_index :
- 1
- product :
- model-output
- project_id :
- EERIE
- realization_index :
- 1
- realm :
- atmos
- references :
- Hohenegger et al., ICON-Sapphire: simulating the components of the Earth system and their interactions at kilometer and subkilometer scales. Geosci. Model Dev., 16, 779–811, 2023, https://doi.org/10.5194/gmd-16-779-2023
- source :
- ICON-ESM-ER (2023): aerosol: none, prescribed MACv2-SP atmos: ICON-A (icosahedral/triangles; 10 km; 90 levels; top level 80 km) atmosChem: none land: JSBACH4.20 landIce: none/prescribed ocean: ICON-O (icosahedral/triangles; 5 km; 72 levels; top grid cell 0-2 m) ocnBgchem: none seaIce: unnamed (thermodynamic (Semtner zero-layer) dynamic (Hibler 79) sea ice model)
- source_id :
- ICON-ESM-ER
- source_type :
- AOGCM
- sub_experiment :
- none
- sub_experiment_id :
- none
- variant_label :
- r1i1p1f1
- version_id :
- v20240618
Environment#
To succesfully run this notebook, you need a kernel / environment with the following packages installed:
- xarray
- pystac
- zarr
- dask
- hvplot # for plotting
- aiohttp
- requests
- intake-xarray
- intake<2
- eccodes # for grib data
- python-eccodes # for grib data
