Example notebook#
Calling the libraries
from getpass import getuser # Libaray to copy things
from pathlib import Path # Object oriented libary to deal with paths
from tempfile import (
NamedTemporaryFile,
TemporaryDirectory,
) # Creating temporary Files/Dirs
from dask.utils import format_bytes
from distributed import (
Client,
progress,
wait,
) # Libaray to orchestrate distributed resources
from dask_jobqueue import SLURMCluster # Setting up distributed memories via slurm
import numpy as np # Pythons standard array library
import xarray as xr # Libary to work with labeled n-dimensional data
import dask # Distributed data libary
import dask.distributed
import multiprocessing
from subprocess import run, PIPE
import sys
import os
import warnings
warnings.filterwarnings(action="ignore")
import pandas as pd
dask.config.config.get("distributed").get("dashboard").update(
{"link": "{JUPYTERHUB_SERVICE_PREFIX}/proxy/{port}/status"}
)
Establish the number of task to work with
ncpu = multiprocessing.cpu_count()
processes = False
nworker = 2
threads = ncpu // nworker
print(
f"Number of CPUs: {ncpu}, number of threads: {threads}, number of workers: {nworker}, processes: {processes}"
)
client = Client(
processes=processes,
threads_per_worker=threads,
n_workers=nworker,
memory_limit="128GB",
)
client
Number of CPUs: 48, number of threads: 24, number of workers: 2, processes: False
Client
|
Cluster
|
Selecting data
# Define a global pattern to find the files
###cloud liquid water
glob_pattern_3d1 = "atm_2d_ml_2020"
## Define the paths of file 1 (dppp0016)
data_path1 = Path("/work/mh0287/k203123/GIT/icon-aes-dyw2/experiments/dpp0029")
## Collect all file names with pathlib's rglob and list compression -- cloud liquid water
file_names1 = sorted([str(f) for f in data_path1.rglob(f"*{glob_pattern_3d1}*.nc")])[:]
Open data set
dset1 = xr.open_mfdataset(
file_names1, combine="by_coords", chunks="auto", engine="netcdf4", parallel=True
)
dset1.info
<bound method Dataset.info of <xarray.Dataset>
Dimensions: (ncells: 20971520, time: 466)
Coordinates:
* time (time) float64 2.02e+07 2.02e+07 2.02e+07 ... 2.02e+07 2.02e+07
Dimensions without coordinates: ncells
Data variables:
ps (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
psl (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsdt (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsut (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsutcs (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rlut (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rlutcs (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsds (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsdscs (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rlds (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rldscs (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsus (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rsuscs (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
rlus (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
ts (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
sic (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
sit (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
clt (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
prlr (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
prls (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
pr (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
prw (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
cllvi (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
clivi (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
qgvi (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
qrvi (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
qsvi (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
hfls (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
hfss (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
evspsbl (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
tauu (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
tauv (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
sfcwind (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
uas (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
vas (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
tas (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
Attributes:
CDI: Climate Data Interface version 1.8.3rc (http://mpim...
Conventions: CF-1.6
number_of_grid_used: 15
grid_file_uri: http://icon-downloads.mpimet.mpg.de/grids/public/mp...
uuidOfHGrid: 0f1e7d66-637e-11e8-913b-51232bb4d8f9
title: ICON simulation
institution: Max Planck Institute for Meteorology/Deutscher Wett...
source: git@gitlab.dkrz.de:icon/icon-aes.git@432d202fb95b6d...
history: /work/mh0287/k203123/GIT/icon-aes-dyw2/bin/icon_dpp...
references: see MPIM/DWD publications
comment: Sapphire Dyamond (k203123) on m21281 (Linux 2.6.32-...>
format_bytes(dset1.nbytes)
'1.41 TB'
Selecting variables
dset_subset = dset1[["ts", "pr"]]
dset_subset.info
<bound method Dataset.info of <xarray.Dataset>
Dimensions: (ncells: 20971520, time: 466)
Coordinates:
* time (time) float64 2.02e+07 2.02e+07 2.02e+07 ... 2.02e+07 2.02e+07
Dimensions without coordinates: ncells
Data variables:
ts (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
pr (time, ncells) float32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
Attributes:
CDI: Climate Data Interface version 1.8.3rc (http://mpim...
Conventions: CF-1.6
number_of_grid_used: 15
grid_file_uri: http://icon-downloads.mpimet.mpg.de/grids/public/mp...
uuidOfHGrid: 0f1e7d66-637e-11e8-913b-51232bb4d8f9
title: ICON simulation
institution: Max Planck Institute for Meteorology/Deutscher Wett...
source: git@gitlab.dkrz.de:icon/icon-aes.git@432d202fb95b6d...
history: /work/mh0287/k203123/GIT/icon-aes-dyw2/bin/icon_dpp...
references: see MPIM/DWD publications
comment: Sapphire Dyamond (k203123) on m21281 (Linux 2.6.32-...>
format_bytes(dset_subset.nbytes)
'78.18 GB'
Changing time
dset_subset.time
<xarray.DataArray 'time' (time: 466)>
array([20200120. , 20200120.25, 20200120.5 , ..., 20201229. , 20201230. ,
20201231. ])
Coordinates:
* time (time) float64 2.02e+07 2.02e+07 2.02e+07 ... 2.02e+07 2.02e+07
Attributes:
standard_name: time
units: day as %Y%m%d.%f
calendar: proleptic_gregorian
axis: Txarray.DataArray
'time'
- time: 466
- 2.02e+07 2.02e+07 2.02e+07 2.02e+07 ... 2.02e+07 2.02e+07 2.02e+07
array([20200120. , 20200120.25, 20200120.5 , ..., 20201229. , 20201230. , 20201231. ]) - time(time)float642.02e+07 2.02e+07 ... 2.02e+07
- standard_name :
- time
- units :
- day as %Y%m%d.%f
- calendar :
- proleptic_gregorian
- axis :
- T
array([20200120. , 20200120.25, 20200120.5 , ..., 20201229. , 20201230. , 20201231. ])
- standard_name :
- time
- units :
- day as %Y%m%d.%f
- calendar :
- proleptic_gregorian
- axis :
- T
time1 = pd.to_datetime(dset_subset.time.data, format="%Y%m%d")
hours = (dset_subset.time.values % 1) * 24
time2 = pd.to_datetime(hours, format="%H")
timenew = pd.to_datetime(pd.to_numeric(time2 - time2[0]) + pd.to_numeric(time1))
dset_subset["time"] = timenew
dset_subset.time
<xarray.DataArray 'time' (time: 466)>
array(['2020-01-20T00:00:00.000000000', '2020-01-20T06:00:00.000000000',
'2020-01-20T12:00:00.000000000', ..., '2020-12-29T00:00:00.000000000',
'2020-12-30T00:00:00.000000000', '2020-12-31T00:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 2020-01-20 2020-01-20T06:00:00 ... 2020-12-31xarray.DataArray
'time'
- time: 466
- 2020-01-20 2020-01-20T06:00:00 ... 2020-12-30 2020-12-31
array(['2020-01-20T00:00:00.000000000', '2020-01-20T06:00:00.000000000', '2020-01-20T12:00:00.000000000', ..., '2020-12-29T00:00:00.000000000', '2020-12-30T00:00:00.000000000', '2020-12-31T00:00:00.000000000'], dtype='datetime64[ns]') - time(time)datetime64[ns]2020-01-20 ... 2020-12-31
array(['2020-01-20T00:00:00.000000000', '2020-01-20T06:00:00.000000000', '2020-01-20T12:00:00.000000000', ..., '2020-12-29T00:00:00.000000000', '2020-12-30T00:00:00.000000000', '2020-12-31T00:00:00.000000000'], dtype='datetime64[ns]')
Daily mean
dset_daily = (
dset_subset.sel(time=slice("2020-02-10", "2020-03-10"))
.resample(time="1D", skipna=True)
.mean()
)
dset_daily
<xarray.Dataset>
Dimensions: (ncells: 20971520, time: 30)
Coordinates:
* time (time) datetime64[ns] 2020-02-10 2020-02-11 ... 2020-03-10
Dimensions without coordinates: ncells
Data variables:
ts (time, ncells) float32 dask.array<chunksize=(1, 5242880), meta=np.ndarray>
pr (time, ncells) float32 dask.array<chunksize=(1, 5242880), meta=np.ndarray>xarray.Dataset
- ncells: 20971520
- time: 30
- time(time)datetime64[ns]2020-02-10 ... 2020-03-10
array(['2020-02-10T00:00:00.000000000', '2020-02-11T00:00:00.000000000', '2020-02-12T00:00:00.000000000', '2020-02-13T00:00:00.000000000', '2020-02-14T00:00:00.000000000', '2020-02-15T00:00:00.000000000', '2020-02-16T00:00:00.000000000', '2020-02-17T00:00:00.000000000', '2020-02-18T00:00:00.000000000', '2020-02-19T00:00:00.000000000', '2020-02-20T00:00:00.000000000', '2020-02-21T00:00:00.000000000', '2020-02-22T00:00:00.000000000', '2020-02-23T00:00:00.000000000', '2020-02-24T00:00:00.000000000', '2020-02-25T00:00:00.000000000', '2020-02-26T00:00:00.000000000', '2020-02-27T00:00:00.000000000', '2020-02-28T00:00:00.000000000', '2020-02-29T00:00:00.000000000', '2020-03-01T00:00:00.000000000', '2020-03-02T00:00:00.000000000', '2020-03-03T00:00:00.000000000', '2020-03-04T00:00:00.000000000', '2020-03-05T00:00:00.000000000', '2020-03-06T00:00:00.000000000', '2020-03-07T00:00:00.000000000', '2020-03-08T00:00:00.000000000', '2020-03-09T00:00:00.000000000', '2020-03-10T00:00:00.000000000'], dtype='datetime64[ns]')
- ts(time, ncells)float32dask.array<chunksize=(1, 5242880), meta=np.ndarray>
Array Chunk Bytes 2.52 GB 20.97 MB Shape (30, 20971520) (1, 5242880) Count 5317 Tasks 120 Chunks Type float32 numpy.ndarray - pr(time, ncells)float32dask.array<chunksize=(1, 5242880), meta=np.ndarray>
Array Chunk Bytes 2.52 GB 20.97 MB Shape (30, 20971520) (1, 5242880) Count 5317 Tasks 120 Chunks Type float32 numpy.ndarray
prmean = dset_daily.pr.mean(dim="time").compute()
format_bytes(prmean.nbytes)
'83.89 MB'
tsmean = dset_daily.ts.mean(dim="time").compute()
format_bytes(tsmean.nbytes)
'83.89 MB'
Assign coordinates
####grid information
filegrid = "/pool/data/ICON/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc"
dsetgrid = xr.open_dataset(filegrid, chunks="auto", engine="netcdf4").rename(
{"cell": "ncells"}
)
dsetgrid
<xarray.Dataset>
Dimensions: (cell_grf: 14, edge: 31457280, edge_grf: 24, max_chdom: 1, max_stored_decompositions: 4, nc: 2, ncells: 20971520, ne: 6, no: 4, nv: 3, two_grf: 2, vert_grf: 13, vertex: 10485762)
Coordinates:
clon (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
clat (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
vlon (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
vlat (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
elon (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
elat (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
Dimensions without coordinates: cell_grf, edge, edge_grf, max_chdom, max_stored_decompositions, nc, ncells, ne, no, nv, two_grf, vert_grf, vertex
Data variables:
clon_vertices (ncells, nv) float64 dask.array<chunksize=(5242880, 3), meta=np.ndarray>
clat_vertices (ncells, nv) float64 dask.array<chunksize=(5242880, 3), meta=np.ndarray>
vlon_vertices (vertex, ne) float64 dask.array<chunksize=(1747627, 6), meta=np.ndarray>
vlat_vertices (vertex, ne) float64 dask.array<chunksize=(1747627, 6), meta=np.ndarray>
elon_vertices (edge, no) float64 dask.array<chunksize=(3932160, 4), meta=np.ndarray>
elat_vertices (edge, no) float64 dask.array<chunksize=(3932160, 4), meta=np.ndarray>
ifs2icon_cell_grid (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
ifs2icon_edge_grid (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
ifs2icon_vertex_grid (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
cell_area (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
dual_area (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
phys_cell_id (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
phys_edge_id (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
lon_cell_centre (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
lat_cell_centre (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
lat_cell_barycenter (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
lon_cell_barycenter (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
longitude_vertices (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
latitude_vertices (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
lon_edge_centre (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
lat_edge_centre (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_of_cell (nv, ncells) int32 dask.array<chunksize=(3, 10485760), meta=np.ndarray>
vertex_of_cell (nv, ncells) int32 dask.array<chunksize=(3, 10485760), meta=np.ndarray>
adjacent_cell_of_edge (nc, edge) int32 dask.array<chunksize=(2, 15728640), meta=np.ndarray>
edge_vertices (nc, edge) int32 dask.array<chunksize=(2, 15728640), meta=np.ndarray>
cells_of_vertex (ne, vertex) int32 dask.array<chunksize=(6, 5242881), meta=np.ndarray>
edges_of_vertex (ne, vertex) int32 dask.array<chunksize=(6, 5242881), meta=np.ndarray>
vertices_of_vertex (ne, vertex) int32 dask.array<chunksize=(6, 5242881), meta=np.ndarray>
cell_area_p (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
cell_elevation (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
cell_sea_land_mask (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
cell_domain_id (ncells, max_stored_decompositions) int32 dask.array<chunksize=(5242880, 4), meta=np.ndarray>
cell_no_of_domains (max_stored_decompositions) int32 dask.array<chunksize=(4,), meta=np.ndarray>
dual_area_p (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
edge_length (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_cell_distance (nc, edge) float64 dask.array<chunksize=(2, 7864320), meta=np.ndarray>
dual_edge_length (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edgequad_area (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_elevation (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_sea_land_mask (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
edge_vert_distance (nc, edge) float64 dask.array<chunksize=(2, 7864320), meta=np.ndarray>
zonal_normal_primal_edge (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
meridional_normal_primal_edge (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
zonal_normal_dual_edge (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
meridional_normal_dual_edge (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
orientation_of_normal (nv, ncells) int32 dask.array<chunksize=(3, 10485760), meta=np.ndarray>
cell_index (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
parent_cell_index (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
parent_cell_type (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
neighbor_cell_index (nv, ncells) int32 dask.array<chunksize=(3, 10485760), meta=np.ndarray>
child_cell_index (no, ncells) int32 dask.array<chunksize=(4, 5242880), meta=np.ndarray>
child_cell_id (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
edge_index (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
edge_parent_type (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
vertex_index (vertex) int32 dask.array<chunksize=(10485762,), meta=np.ndarray>
edge_orientation (ne, vertex) int32 dask.array<chunksize=(6, 5242881), meta=np.ndarray>
edge_system_orientation (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
refin_c_ctrl (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
index_c_list (two_grf, cell_grf) int32 dask.array<chunksize=(2, 14), meta=np.ndarray>
start_idx_c (max_chdom, cell_grf) int32 dask.array<chunksize=(1, 14), meta=np.ndarray>
end_idx_c (max_chdom, cell_grf) int32 dask.array<chunksize=(1, 14), meta=np.ndarray>
refin_e_ctrl (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
index_e_list (two_grf, edge_grf) int32 dask.array<chunksize=(2, 24), meta=np.ndarray>
start_idx_e (max_chdom, edge_grf) int32 dask.array<chunksize=(1, 24), meta=np.ndarray>
end_idx_e (max_chdom, edge_grf) int32 dask.array<chunksize=(1, 24), meta=np.ndarray>
refin_v_ctrl (vertex) int32 dask.array<chunksize=(10485762,), meta=np.ndarray>
index_v_list (two_grf, vert_grf) int32 dask.array<chunksize=(2, 13), meta=np.ndarray>
start_idx_v (max_chdom, vert_grf) int32 dask.array<chunksize=(1, 13), meta=np.ndarray>
end_idx_v (max_chdom, vert_grf) int32 dask.array<chunksize=(1, 13), meta=np.ndarray>
parent_edge_index (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
child_edge_index (no, edge) int32 dask.array<chunksize=(4, 7864320), meta=np.ndarray>
child_edge_id (edge) int32 dask.array<chunksize=(31457280,), meta=np.ndarray>
parent_vertex_index (vertex) int32 dask.array<chunksize=(10485762,), meta=np.ndarray>
cartesian_x_vertices (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
cartesian_y_vertices (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
cartesian_z_vertices (vertex) float64 dask.array<chunksize=(10485762,), meta=np.ndarray>
edge_middle_cartesian_x (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_middle_cartesian_y (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_middle_cartesian_z (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_dual_middle_cartesian_x (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_dual_middle_cartesian_y (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_dual_middle_cartesian_z (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_primal_normal_cartesian_x (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_primal_normal_cartesian_y (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_primal_normal_cartesian_z (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_dual_normal_cartesian_x (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_dual_normal_cartesian_y (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
edge_dual_normal_cartesian_z (edge) float64 dask.array<chunksize=(15728640,), meta=np.ndarray>
cell_circumcenter_cartesian_x (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
cell_circumcenter_cartesian_y (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
cell_circumcenter_cartesian_z (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
Attributes:
title: ICON grid description
institution: Max Planck Institute for Meteorology/Deutscher ...
source: git@git.mpimet.mpg.de:GridGenerator.git
revision: d00fcac1f61fa16c686bfe51d1d8eddd09296cb5
date: 20180529 at 222250
user_name: Rene Redler (m300083)
os_name: Linux 2.6.32-696.18.7.el6.x86_64 x86_64
uuidOfHGrid: 0f1e7d66-637e-11e8-913b-51232bb4d8f9
grid_mapping_name: lat_long_on_sphere
crs_id: urn:ogc:def:cs:EPSG:6.0:6422
crs_name: Spherical 2D Coordinate System
ellipsoid_name: Sphere
semi_major_axis: 6371229.0
inverse_flattening: 0.0
grid_level: 9
grid_root: 2
grid_ID: 1
parent_grid_ID: 0
no_of_subgrids: 1
start_subgrid_id: 1
max_childdom: 1
boundary_depth_index: 0
rotation_vector: [0. 0. 0.]
grid_geometry: 1
grid_cell_type: 3
mean_edge_length: 7510.64679407352
mean_dual_edge_length: 4336.344345177032
mean_cell_area: 24323517.809282698
mean_dual_cell_area: 48647026.33989711
domain_length: 40031612.44147649
domain_height: 40031612.44147649
sphere_radius: 6371229.0
domain_cartesian_center: [0. 0. 0.]
centre: 252
rotation: 37deg around z-axis
coverage: global
symmetry: along equator
topography: modified SRTM30
subcentre: 1
number_of_grid_used: 15
history: Thu Aug 16 11:05:44 2018: ncatted -O -a ICON_gr...
ICON_grid_file_uri: http://icon-downloads.mpimet.mpg.de/grids/publi...
NCO: netCDF Operators version 4.7.5 (Homepage = http...xarray.Dataset
- cell_grf: 14
- edge: 31457280
- edge_grf: 24
- max_chdom: 1
- max_stored_decompositions: 4
- nc: 2
- ncells: 20971520
- ne: 6
- no: 4
- nv: 3
- two_grf: 2
- vert_grf: 13
- vertex: 10485762
- clon(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- center longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- clon_vertices
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - clat(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- center latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- clat_vertices
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - vlon(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- vertex longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- vlon_vertices
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - vlat(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- vertex latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- vlat_vertices
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - elon(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- edge midpoint longitude
- units :
- radian
- standard_name :
- grid_longitude
- bounds :
- elon_vertices
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - elat(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- edge midpoint latitude
- units :
- radian
- standard_name :
- grid_latitude
- bounds :
- elat_vertices
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray
- clon_vertices(ncells, nv)float64dask.array<chunksize=(5242880, 3), meta=np.ndarray>
- units :
- radian
Array Chunk Bytes 503.32 MB 125.83 MB Shape (20971520, 3) (5242880, 3) Count 5 Tasks 4 Chunks Type float64 numpy.ndarray - clat_vertices(ncells, nv)float64dask.array<chunksize=(5242880, 3), meta=np.ndarray>
- units :
- radian
Array Chunk Bytes 503.32 MB 125.83 MB Shape (20971520, 3) (5242880, 3) Count 5 Tasks 4 Chunks Type float64 numpy.ndarray - vlon_vertices(vertex, ne)float64dask.array<chunksize=(1747627, 6), meta=np.ndarray>
- units :
- radian
Array Chunk Bytes 503.32 MB 83.89 MB Shape (10485762, 6) (1747627, 6) Count 7 Tasks 6 Chunks Type float64 numpy.ndarray - vlat_vertices(vertex, ne)float64dask.array<chunksize=(1747627, 6), meta=np.ndarray>
- units :
- radian
Array Chunk Bytes 503.32 MB 83.89 MB Shape (10485762, 6) (1747627, 6) Count 7 Tasks 6 Chunks Type float64 numpy.ndarray - elon_vertices(edge, no)float64dask.array<chunksize=(3932160, 4), meta=np.ndarray>
- units :
- radian
Array Chunk Bytes 1.01 GB 125.83 MB Shape (31457280, 4) (3932160, 4) Count 9 Tasks 8 Chunks Type float64 numpy.ndarray - elat_vertices(edge, no)float64dask.array<chunksize=(3932160, 4), meta=np.ndarray>
- units :
- radian
Array Chunk Bytes 1.01 GB 125.83 MB Shape (31457280, 4) (3932160, 4) Count 9 Tasks 8 Chunks Type float64 numpy.ndarray - ifs2icon_cell_grid(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- ifs to icon cells
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - ifs2icon_edge_grid(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- ifs to icon edge
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - ifs2icon_vertex_grid(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- ifs to icon vertex
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - cell_area(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- area of grid cell
- units :
- m2
- standard_name :
- area
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - dual_area(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- areas of dual hexagonal/pentagonal cells
- units :
- m2
- standard_name :
- area
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - phys_cell_id(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- physical domain ID of cell
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - phys_edge_id(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- physical domain ID of edge
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - lon_cell_centre(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- longitude of cell centre
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - lat_cell_centre(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- latitude of cell centre
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - lat_cell_barycenter(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- latitude of cell barycenter
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - lon_cell_barycenter(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- longitude of cell barycenter
- units :
- radian
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - longitude_vertices(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- longitude of vertices
- units :
- radian
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - latitude_vertices(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- latitude of vertices
- units :
- radian
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lon_edge_centre(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- longitudes of edge midpoints
- units :
- radian
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - lat_edge_centre(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- latitudes of edge midpoints
- units :
- radian
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_of_cell(nv, ncells)int32dask.array<chunksize=(3, 10485760), meta=np.ndarray>
- long_name :
- edges of each cellvertices
Array Chunk Bytes 251.66 MB 125.83 MB Shape (3, 20971520) (3, 10485760) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - vertex_of_cell(nv, ncells)int32dask.array<chunksize=(3, 10485760), meta=np.ndarray>
- long_name :
- vertices of each cellcells ad
Array Chunk Bytes 251.66 MB 125.83 MB Shape (3, 20971520) (3, 10485760) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - adjacent_cell_of_edge(nc, edge)int32dask.array<chunksize=(2, 15728640), meta=np.ndarray>
- long_name :
- cells adjacent to each edge
Array Chunk Bytes 251.66 MB 125.83 MB Shape (2, 31457280) (2, 15728640) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - edge_vertices(nc, edge)int32dask.array<chunksize=(2, 15728640), meta=np.ndarray>
- long_name :
- vertices at the end of of each edge
Array Chunk Bytes 251.66 MB 125.83 MB Shape (2, 31457280) (2, 15728640) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - cells_of_vertex(ne, vertex)int32dask.array<chunksize=(6, 5242881), meta=np.ndarray>
- long_name :
- cells around each vertex
Array Chunk Bytes 251.66 MB 125.83 MB Shape (6, 10485762) (6, 5242881) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - edges_of_vertex(ne, vertex)int32dask.array<chunksize=(6, 5242881), meta=np.ndarray>
- long_name :
- edges around each vertex
Array Chunk Bytes 251.66 MB 125.83 MB Shape (6, 10485762) (6, 5242881) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - vertices_of_vertex(ne, vertex)int32dask.array<chunksize=(6, 5242881), meta=np.ndarray>
- long_name :
- vertices around each vertex
Array Chunk Bytes 251.66 MB 125.83 MB Shape (6, 10485762) (6, 5242881) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - cell_area_p(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- area of grid cell
- units :
- m2
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - cell_elevation(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- elevation at the cell centers
- units :
- m
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - cell_sea_land_mask(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell
- units :
- 2,1,-1,-
- grid_type :
- unstructured
- number_of_grid_in_reference :
- 1
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - cell_domain_id(ncells, max_stored_decompositions)int32dask.array<chunksize=(5242880, 4), meta=np.ndarray>
- long_name :
- cell domain id for decomposition
Array Chunk Bytes 335.54 MB 83.89 MB Shape (20971520, 4) (5242880, 4) Count 5 Tasks 4 Chunks Type int32 numpy.ndarray - cell_no_of_domains(max_stored_decompositions)int32dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- number of domains for each decomposition
Array Chunk Bytes 16 B 16 B Shape (4,) (4,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - dual_area_p(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- areas of dual hexagonal/pentagonal cells
- units :
- m2
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - edge_length(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- lengths of edges of triangular cells
- units :
- m
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_cell_distance(nc, edge)float64dask.array<chunksize=(2, 7864320), meta=np.ndarray>
- long_name :
- distances between edge midpoint and adjacent triangle midpoints
- units :
- m
Array Chunk Bytes 503.32 MB 125.83 MB Shape (2, 31457280) (2, 7864320) Count 5 Tasks 4 Chunks Type float64 numpy.ndarray - dual_edge_length(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- lengths of dual edges (distances between triangular cell circumcenters)
- units :
- m
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edgequad_area(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- area around the edge formed by the two adjacent triangles
- units :
- m2
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_elevation(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- elevation at the edge centers
- units :
- m
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_sea_land_mask(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell
- units :
- 2,1,-1,-
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - edge_vert_distance(nc, edge)float64dask.array<chunksize=(2, 7864320), meta=np.ndarray>
- long_name :
- distances between edge midpoint and vertices of that edge
- units :
- m
Array Chunk Bytes 503.32 MB 125.83 MB Shape (2, 31457280) (2, 7864320) Count 5 Tasks 4 Chunks Type float64 numpy.ndarray - zonal_normal_primal_edge(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- zonal component of normal to primal edge
- units :
- radian
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - meridional_normal_primal_edge(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- meridional component of normal to primal edge
- units :
- radian
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - zonal_normal_dual_edge(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- zonal component of normal to dual edge
- units :
- radian
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - meridional_normal_dual_edge(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- meridional component of normal to dual edge
- units :
- radian
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - orientation_of_normal(nv, ncells)int32dask.array<chunksize=(3, 10485760), meta=np.ndarray>
- long_name :
- orientations of normals to triangular cell edges
Array Chunk Bytes 251.66 MB 125.83 MB Shape (3, 20971520) (3, 10485760) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - cell_index(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- cell index
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - parent_cell_index(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- parent cell index
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - parent_cell_type(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- parent cell type
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - neighbor_cell_index(nv, ncells)int32dask.array<chunksize=(3, 10485760), meta=np.ndarray>
- long_name :
- cell neighbor index
Array Chunk Bytes 251.66 MB 125.83 MB Shape (3, 20971520) (3, 10485760) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - child_cell_index(no, ncells)int32dask.array<chunksize=(4, 5242880), meta=np.ndarray>
- long_name :
- child cell index
Array Chunk Bytes 335.54 MB 83.89 MB Shape (4, 20971520) (4, 5242880) Count 5 Tasks 4 Chunks Type int32 numpy.ndarray - child_cell_id(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- domain ID of child cell
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - edge_index(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- edge index
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - edge_parent_type(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- edge paren
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - vertex_index(vertex)int32dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- vertices index
Array Chunk Bytes 41.94 MB 41.94 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - edge_orientation(ne, vertex)int32dask.array<chunksize=(6, 5242881), meta=np.ndarray>
- long_name :
- edge orientation
Array Chunk Bytes 251.66 MB 125.83 MB Shape (6, 10485762) (6, 5242881) Count 3 Tasks 2 Chunks Type int32 numpy.ndarray - edge_system_orientation(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- edge system orientation
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - refin_c_ctrl(ncells)int32dask.array<chunksize=(20971520,), meta=np.ndarray>
- long_name :
- refinement control flag for cells
Array Chunk Bytes 83.89 MB 83.89 MB Shape (20971520,) (20971520,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - index_c_list(two_grf, cell_grf)int32dask.array<chunksize=(2, 14), meta=np.ndarray>
- long_name :
- list of start and end indices for each refinement control level for cells
Array Chunk Bytes 112 B 112 B Shape (2, 14) (2, 14) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - start_idx_c(max_chdom, cell_grf)int32dask.array<chunksize=(1, 14), meta=np.ndarray>
- long_name :
- list of start indices for each refinement control level for cells
Array Chunk Bytes 56 B 56 B Shape (1, 14) (1, 14) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - end_idx_c(max_chdom, cell_grf)int32dask.array<chunksize=(1, 14), meta=np.ndarray>
- long_name :
- list of end indices for each refinement control level for cells
Array Chunk Bytes 56 B 56 B Shape (1, 14) (1, 14) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - refin_e_ctrl(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- refinement control flag for edges
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - index_e_list(two_grf, edge_grf)int32dask.array<chunksize=(2, 24), meta=np.ndarray>
- long_name :
- list of start and end indices for each refinement control level for edges
Array Chunk Bytes 192 B 192 B Shape (2, 24) (2, 24) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - start_idx_e(max_chdom, edge_grf)int32dask.array<chunksize=(1, 24), meta=np.ndarray>
- long_name :
- list of start indices for each refinement control level for edges
Array Chunk Bytes 96 B 96 B Shape (1, 24) (1, 24) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - end_idx_e(max_chdom, edge_grf)int32dask.array<chunksize=(1, 24), meta=np.ndarray>
- long_name :
- list of end indices for each refinement control level for edges
Array Chunk Bytes 96 B 96 B Shape (1, 24) (1, 24) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - refin_v_ctrl(vertex)int32dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- refinement control flag for vertices
Array Chunk Bytes 41.94 MB 41.94 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - index_v_list(two_grf, vert_grf)int32dask.array<chunksize=(2, 13), meta=np.ndarray>
- long_name :
- list of start and end indices for each refinement control level for vertices
Array Chunk Bytes 104 B 104 B Shape (2, 13) (2, 13) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - start_idx_v(max_chdom, vert_grf)int32dask.array<chunksize=(1, 13), meta=np.ndarray>
- long_name :
- list of start indices for each refinement control level for vertices
Array Chunk Bytes 52 B 52 B Shape (1, 13) (1, 13) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - end_idx_v(max_chdom, vert_grf)int32dask.array<chunksize=(1, 13), meta=np.ndarray>
- long_name :
- list of end indices for each refinement control level for vertices
Array Chunk Bytes 52 B 52 B Shape (1, 13) (1, 13) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - parent_edge_index(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- parent edge index
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - child_edge_index(no, edge)int32dask.array<chunksize=(4, 7864320), meta=np.ndarray>
- long_name :
- child edge index
Array Chunk Bytes 503.32 MB 125.83 MB Shape (4, 31457280) (4, 7864320) Count 5 Tasks 4 Chunks Type int32 numpy.ndarray - child_edge_id(edge)int32dask.array<chunksize=(31457280,), meta=np.ndarray>
- long_name :
- domain ID of child edge
Array Chunk Bytes 125.83 MB 125.83 MB Shape (31457280,) (31457280,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - parent_vertex_index(vertex)int32dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- parent vertex index
Array Chunk Bytes 41.94 MB 41.94 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type int32 numpy.ndarray - cartesian_x_vertices(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- vertex cartesian coordinate x on unit sp
- units :
- meters
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - cartesian_y_vertices(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- vertex cartesian coordinate y on unit sp
- units :
- meters
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - cartesian_z_vertices(vertex)float64dask.array<chunksize=(10485762,), meta=np.ndarray>
- long_name :
- vertex cartesian coordinate z on unit sp
- units :
- meters
Array Chunk Bytes 83.89 MB 83.89 MB Shape (10485762,) (10485762,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - edge_middle_cartesian_x(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- prime edge center cartesian coordinate x on unit sphere
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_middle_cartesian_y(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- prime edge center cartesian coordinate y on unit sphere
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_middle_cartesian_z(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- prime edge center cartesian coordinate z on unit sphere
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_dual_middle_cartesian_x(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- dual edge center cartesian coordinate x on unit sphere
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_dual_middle_cartesian_y(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- dual edge center cartesian coordinate y on unit sphere
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_dual_middle_cartesian_z(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- dual edge center cartesian coordinate z on unit sphere
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_primal_normal_cartesian_x(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- unit normal to the prime edge 3D vector, coordinate x
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_primal_normal_cartesian_y(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- unit normal to the prime edge 3D vector, coordinate y
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_primal_normal_cartesian_z(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- unit normal to the prime edge 3D vector, coordinate z
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_dual_normal_cartesian_x(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- unit normal to the dual edge 3D vector, coordinate x
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_dual_normal_cartesian_y(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- unit normal to the dual edge 3D vector, coordinate y
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - edge_dual_normal_cartesian_z(edge)float64dask.array<chunksize=(15728640,), meta=np.ndarray>
- long_name :
- unit normal to the dual edge 3D vector, coordinate z
- units :
- meters
Array Chunk Bytes 251.66 MB 125.83 MB Shape (31457280,) (15728640,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - cell_circumcenter_cartesian_x(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- cartesian position of the prime cell circumcenter on the unit sphere, coordinate x
- units :
- meters
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - cell_circumcenter_cartesian_y(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- cartesian position of the prime cell circumcenter on the unit sphere, coordinate y
- units :
- meters
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray - cell_circumcenter_cartesian_z(ncells)float64dask.array<chunksize=(10485760,), meta=np.ndarray>
- long_name :
- cartesian position of the prime cell circumcenter on the unit sphere, coordinate z
- units :
- meters
Array Chunk Bytes 167.77 MB 83.89 MB Shape (20971520,) (10485760,) Count 3 Tasks 2 Chunks Type float64 numpy.ndarray
- title :
- ICON grid description
- institution :
- Max Planck Institute for Meteorology/Deutscher Wetterdienst
- source :
- git@git.mpimet.mpg.de:GridGenerator.git
- revision :
- d00fcac1f61fa16c686bfe51d1d8eddd09296cb5
- date :
- 20180529 at 222250
- user_name :
- Rene Redler (m300083)
- os_name :
- Linux 2.6.32-696.18.7.el6.x86_64 x86_64
- uuidOfHGrid :
- 0f1e7d66-637e-11e8-913b-51232bb4d8f9
- grid_mapping_name :
- lat_long_on_sphere
- crs_id :
- urn:ogc:def:cs:EPSG:6.0:6422
- crs_name :
- Spherical 2D Coordinate System
- ellipsoid_name :
- Sphere
- semi_major_axis :
- 6371229.0
- inverse_flattening :
- 0.0
- grid_level :
- 9
- grid_root :
- 2
- grid_ID :
- 1
- parent_grid_ID :
- 0
- no_of_subgrids :
- 1
- start_subgrid_id :
- 1
- max_childdom :
- 1
- boundary_depth_index :
- 0
- rotation_vector :
- [0. 0. 0.]
- grid_geometry :
- 1
- grid_cell_type :
- 3
- mean_edge_length :
- 7510.64679407352
- mean_dual_edge_length :
- 4336.344345177032
- mean_cell_area :
- 24323517.809282698
- mean_dual_cell_area :
- 48647026.33989711
- domain_length :
- 40031612.44147649
- domain_height :
- 40031612.44147649
- sphere_radius :
- 6371229.0
- domain_cartesian_center :
- [0. 0. 0.]
- centre :
- 252
- rotation :
- 37deg around z-axis
- coverage :
- global
- symmetry :
- along equator
- topography :
- modified SRTM30
- subcentre :
- 1
- number_of_grid_used :
- 15
- history :
- Thu Aug 16 11:05:44 2018: ncatted -O -a ICON_grid_file_uri,global,m,c,http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc icon_grid_0015_R02B09_G.nc test.nc Wed May 30 08:50:27 2018: ncatted -a centre,global,c,i,252 -a rotation,global,c,c,37deg around z-axis -a coverage,global,c,c,global -a symmetry,global,c,c,along equator -a topography,global,c,c,modified SRTM30 -a subcentre,global,c,i,1 -a number_of_grid_used,global,c,i,15 -a ICON_grid_file_uri,global,c,c,http://icon-downloads.mpimet.mpg.de/grids/public/icon_grid_0015_R02B09_G.nc Earth_Global_IcosSymmetric_4932m_rotatedZ37d_modified_srtm30_1min.nc icon_grid_0015_R02B09_G.nc /mnt/lustre01/work/mh0287/users/rene/GridGenerator/build/x86_64-unknown-linux-gnu/bin/grid_command
- ICON_grid_file_uri :
- http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
- NCO :
- netCDF Operators version 4.7.5 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
Assing lat and lot to the output file
prmean = prmean.assign_coords(
clon=("ncells", dsetgrid.clon * 180 / np.pi),
clat=("ncells", dsetgrid.clat * 180 / np.pi),
)
prmean.clon
<xarray.DataArray 'clon' (ncells: 20971520)>
array([73. , 73.05645347, 72.94354653, ..., 76.49297292,
76.54481611, 76.4503828 ])
Coordinates:
clon (ncells) float64 73.0 73.06 72.94 73.0 ... 76.5 76.49 76.54 76.45
clat (ncells) float64 52.61 52.63 52.63 52.57 ... -45.17 -45.23 -45.23
Dimensions without coordinates: ncellsxarray.DataArray
'clon'
- ncells: 20971520
- 73.0 73.06 72.94 73.0 73.0 73.06 ... 76.44 76.5 76.49 76.54 76.45
array([73. , 73.05645347, 72.94354653, ..., 76.49297292, 76.54481611, 76.4503828 ]) - clon(ncells)float6473.0 73.06 72.94 ... 76.54 76.45
array([73. , 73.05645347, 72.94354653, ..., 76.49297292, 76.54481611, 76.4503828 ]) - clat(ncells)float6452.61 52.63 52.63 ... -45.23 -45.23
array([ 52.60689104, 52.62663022, 52.62663022, ..., -45.17171677, -45.23138792, -45.23383705])
tsmean = prmean.assign_coords(
clon=("ncells", dsetgrid.clon * 180 / np.pi),
clat=("ncells", dsetgrid.clat * 180 / np.pi),
)
tsmean.clat
<xarray.DataArray 'clat' (ncells: 20971520)>
array([ 52.60689104, 52.62663022, 52.62663022, ..., -45.17171677,
-45.23138792, -45.23383705])
Coordinates:
clon (ncells) float64 73.0 73.06 72.94 73.0 ... 76.5 76.49 76.54 76.45
clat (ncells) float64 52.61 52.63 52.63 52.57 ... -45.17 -45.23 -45.23
Dimensions without coordinates: ncellsxarray.DataArray
'clat'
- ncells: 20971520
- 52.61 52.63 52.63 52.57 52.69 ... -45.11 -45.21 -45.17 -45.23 -45.23
array([ 52.60689104, 52.62663022, 52.62663022, ..., -45.17171677, -45.23138792, -45.23383705]) - clon(ncells)float6473.0 73.06 72.94 ... 76.54 76.45
array([73. , 73.05645347, 72.94354653, ..., 76.49297292, 76.54481611, 76.4503828 ]) - clat(ncells)float6452.61 52.63 52.63 ... -45.23 -45.23
array([ 52.60689104, 52.62663022, 52.62663022, ..., -45.17171677, -45.23138792, -45.23383705])
Plotting
from cartopy import crs as ccrs # Cartography library
from cartopy.mpl.ticker import LongitudeFormatter, LatitudeFormatter
import cartopy.feature as cfeature
from matplotlib.ticker import MultipleLocator, FormatStrFormatter, AutoMinorLocator
from matplotlib import pyplot as plt # Standard Plotting library
from matplotlib import cm
from matplotlib.colors import ListedColormap, LinearSegmentedColormap
Using native grid
fig = plt.figure(figsize=(10, 6))
ax = fig.add_subplot(111, projection=ccrs.PlateCarree(central_longitude=0))
cs1 = plt.scatter(
prmean.clon[::100],
prmean.clat[::100],
s=0.1,
c=prmean[::100] * 24 * 3600,
vmin=0,
vmax=14,
cmap="YlGnBu",
transform=ccrs.PlateCarree(),
)
cb1 = plt.colorbar(cs1, extend="max", orientation="horizontal", shrink=0.6, pad=0.1)
cb1.ax.set_xlabel("Precipitation (mm day$^{-1}$)", fontsize=12)
cb1.ax.tick_params(labelsize=12)
ax.coastlines(resolution="110m", linewidth=0.6)
ax.set_extent([-180, 179.9, -80, 80], crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(-80, 81, 20), crs=ccrs.PlateCarree())
lat_formatter = LatitudeFormatter()
ax.yaxis.set_major_formatter(lat_formatter)
ax.set_xticks(np.arange(-180, 179, 60), crs=ccrs.PlateCarree())
lon_formatter = LongitudeFormatter(zero_direction_label=True)
ax.xaxis.set_major_formatter(lon_formatter)
ax.tick_params(labelsize=12)
ax.set_xlabel(None)
ax.set_ylabel(None)
Text(0, 0.5, '')
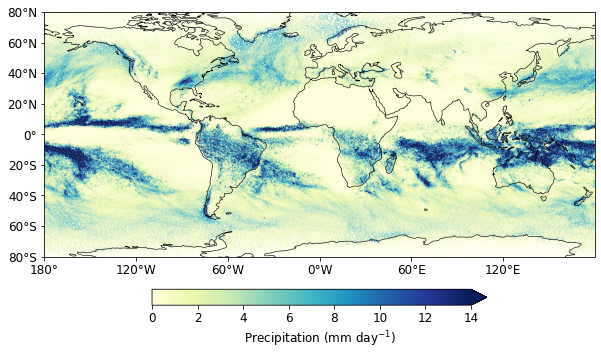
Assign land_sea mask
land_sea_mask = dsetgrid.cell_sea_land_mask.persist()
prmean["land_sea_mask"] = (("ncells"), land_sea_mask)
prmean_land = prmean.where(prmean.land_sea_mask > 0)
fig = plt.figure(figsize=(10, 6))
ax = fig.add_subplot(111, projection=ccrs.PlateCarree(central_longitude=0))
cs1 = plt.scatter(
prmean_land.clon[::100],
prmean_land.clat[::100],
s=0.1,
c=prmean_land[::100] * 24 * 3600,
vmin=0,
vmax=14,
cmap="YlGnBu",
transform=ccrs.PlateCarree(),
)
cb1 = plt.colorbar(cs1, extend="max", orientation="horizontal", shrink=0.6, pad=0.1)
cb1.ax.set_xlabel("Precipitation (mm day$^{-1}$)", fontsize=12)
cb1.ax.tick_params(labelsize=12)
ax.coastlines(resolution="110m", linewidth=0.6)
ax.set_extent([-180, 179.9, -80, 80], crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(-80, 81, 20), crs=ccrs.PlateCarree())
lat_formatter = LatitudeFormatter()
ax.yaxis.set_major_formatter(lat_formatter)
ax.set_xticks(np.arange(-180, 179, 60), crs=ccrs.PlateCarree())
lon_formatter = LongitudeFormatter(zero_direction_label=True)
ax.xaxis.set_major_formatter(lon_formatter)
ax.tick_params(labelsize=12)
ax.set_xlabel(None)
ax.set_ylabel(None)
Text(0, 0.5, '')
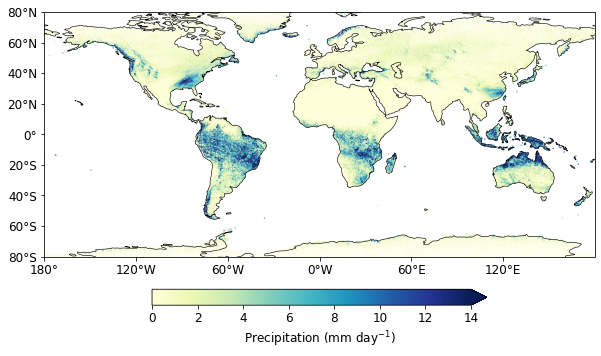
Regridding
def get_griddes(y_res, x_res, x_first=-180, y_first=-90):
"""Create a description for a regular global grid at given x, y resolution."""
xsize = 360 / x_res
ysize = 180 / y_res
xfirst = -180 + x_res / 2
yfirst = -90 + x_res / 2
return f"""
#
# gridID 1
#
gridtype = lonlat
gridsize = {int(xsize * ysize)}
xsize = {int(xsize)}
ysize = {int(ysize)}
xname = lon
xlongname = "longitude"
xunits = "degrees_east"
yname = lat
ylongname = "latitude"
yunits = "degrees_north"
xfirst = {xfirst}
xinc = {x_res}
yfirst = {yfirst}
yinc = {y_res}
"""
@dask.delayed
def gen_dis(dataset, xres, yres, gridfile):
"""Create a distance weights using cdo."""
scratch_dir = (
Path("/scratch") / getuser()[0] / getuser() / Path("data")
) # Define the users scratch dir
with TemporaryDirectory(dir=scratch_dir, prefix="Weights_") as td:
in_file = Path(td) / "in_file.nc"
weightfile = Path(td) / "weight_file.nc"
griddes = Path(td) / "griddes.txt"
with griddes.open("w") as f:
f.write(get_griddes(xres, yres))
dataset.to_netcdf(in_file, mode="w") # Write the file to a temorary netcdf file
cmd = (
"cdo",
"-O",
f"gendis,{griddes}",
f"-setgrid,{gridfile}",
str(in_file),
str(weightfile),
)
run_cmd(cmd)
df = xr.open_dataset(weightfile).load()
wait(df)
return df
def run_cmd(cmd, path_extra=Path(sys.exec_prefix) / "bin"):
"""Run a bash command."""
env_extra = os.environ.copy()
env_extra["PATH"] = str(path_extra) + ":" + env_extra["PATH"]
status = run(cmd, check=False, stderr=PIPE, stdout=PIPE, env=env_extra)
if status.returncode != 0:
error = f"""{' '.join(cmd)}: {status.stderr.decode('utf-8')}"""
raise RuntimeError(f"{error}")
return status.stdout.decode("utf-8")
@dask.delayed
def remap(dataset, x_res, y_res, weights, gridfile):
"""Perform a weighted remapping.
Parameters
==========
dataset : xarray.dataset
The dataset that will be regridded
griddes : Path, str
Path to the grid description file
weights : xarray.dataset
Distance weights
Returns
=======
xarray.dataset : Remapped dataset
"""
if isinstance(dataset, xr.DataArray):
# If a dataArray is given create a dataset
dataset = xr.Dataset(data_vars={dataset.name: dataset})
scratch_dir = (
Path("/scratch") / getuser()[0] / getuser() / Path("data")
) # Define the users scratch dir
with TemporaryDirectory(dir=scratch_dir, prefix="Remap_") as td:
infile = Path(td) / "input_file.nc"
weightfile = Path(td) / "weight_file.nc"
griddes = Path(td) / "griddes.txt"
outfile = Path(td) / "remaped_file.nc"
with griddes.open("w") as f:
f.write(get_griddes(x_res, y_res))
dataset.to_netcdf(infile, mode="w") # Write the file to a temorary netcdf file
weights.to_netcdf(weightfile, mode="w")
cmd = (
"cdo",
"-O",
f"remap,{griddes},{weightfile}",
f"-setgrid,{gridfile}",
str(infile),
str(outfile),
)
run_cmd(cmd)
return xr.open_dataset(outfile).load()
Create weighted grids
weights_future = gen_dis(prmean, 0.1, 0.1, filegrid)
Remapping grids
####File 1
remap_precip = remap(prmean, 0.1, 0.1, weights_future, filegrid)
remap_jobs = dask.persist(remap_precip)
progress(remap_jobs, notebook=False)
[########################################] | 100% Completed | 45.6s
precip = dask.compute(remap_precip)[0]
precip.info
<bound method Dataset.info of <xarray.Dataset>
Dimensions: (lat: 1800, lon: 3600)
Coordinates:
* lon (lon) float64 -179.9 -179.8 -179.8 -179.6 ... 179.8 179.9 180.0
* lat (lat) float64 -89.95 -89.85 -89.75 -89.65 ... 89.75 89.85 89.95
Data variables:
pr (lat, lon) float32 1.695e-06 1.695e-06 ... 5.842e-06 5.842e-06
Attributes:
CDI: Climate Data Interface version 1.9.8 (https://mpimet.mpg.de...
Conventions: CF-1.6
history: Wed Sep 08 11:02:08 2021: cdo -O remap,/scratch/m/m300876/d...
CDO: Climate Data Operators version 1.9.8 (https://mpimet.mpg.de...>
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(111, projection=ccrs.PlateCarree(central_longitude=225))
plot = (precip.pr * 24 * 3600).plot(
ax=ax,
transform=ccrs.PlateCarree(),
cmap="YlGnBu",
levels=np.linspace(0, 14, 15),
vmin=0,
vmax=14,
extend="both",
add_colorbar=False,
)
cb1 = plt.colorbar(plot, extend="max", orientation="horizontal", shrink=0.8, pad=0.12)
cb1.ax.set_xlabel("Altitude (Km)", fontsize=12)
cb1.ax.tick_params(labelsize=12)
ax.coastlines(resolution="110m", linewidth=0.6)
ax.set_extent([-180, 179.9, -80, 80], crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(-80, 81, 20), crs=ccrs.PlateCarree())
lat_formatter = LatitudeFormatter()
ax.yaxis.set_major_formatter(lat_formatter)
ax.set_xticks(np.arange(-180, 179, 60), crs=ccrs.PlateCarree())
lon_formatter = LongitudeFormatter(zero_direction_label=True)
ax.xaxis.set_major_formatter(lon_formatter)
ax.tick_params(labelsize=12)
ax.set_xlabel(None)
ax.set_ylabel(None)
Text(0, 0.5, '')
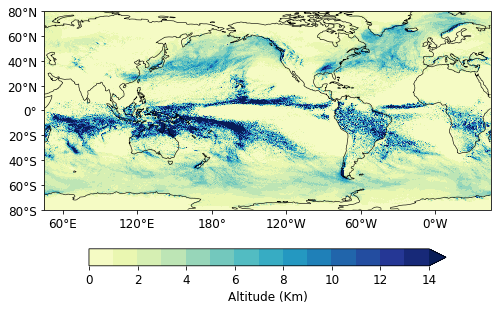
Save file
scratch_dir = (
Path("/scratch") / getuser()[0] / getuser()
) # if it has not been defined before
out_file = Path(scratch_dir) / "OutfileName.nc"
prmean.to_netcdf(out_file, mode="w")
Load file
loadset = xr.open_dataset(out_file, chunks="auto", engine="netcdf4")
loadset.info
<bound method Dataset.info of <xarray.Dataset>
Dimensions: (ncells: 20971520)
Coordinates:
clon (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
clat (ncells) float64 dask.array<chunksize=(10485760,), meta=np.ndarray>
land_sea_mask (ncells) int32 dask.array<chunksize=(20971520,), meta=np.ndarray>
Dimensions without coordinates: ncells
Data variables:
pr (ncells) float32 dask.array<chunksize=(20971520,), meta=np.ndarray>>
