Working with Dask#
Earth system datasets are often too large to fit into memory, and analyses frequently need to scale from a laptop to a compute cluster. Dask addresses this challenge by extending Python tools such as xarray to work lazily and in parallel. Instead of executing operations immediately, Dask builds a task graph that describes what needs to be done and only performs the actual computation when explicitly requested. This design allows users to prototype analyses interactively while retaining the option to scale up without rewriting code.
At the user level, Dask is primarily encountered through its collections such as dask.array, dask.dataframe, and xarray objects backed by Dask.
These collections look and feel like their in-memory counterparts but are internally split into smaller chunks.
Chunking is a key concept: it determines how data is partitioned and therefore strongly influences performance and memory usage.
In practice, users should think less about Dask internals and more about choosing chunk sizes that align with their access patterns (e.g. spatial vs. temporal operations in climate data) and the available compute resources.
Finally, Dask separates defining a computation from executing it.
Operations on Dask-backed objects are cheap and immediate, while execution happens only when calling methods such as .compute() or when results are plotted or written to disk.
This separation enables parallel execution on a single machine or a cluster via a scheduler, without changing the analysis logic.
For Earth system science workflows, this means Dask can serve as a scalable execution engine underneath xarray-based analyses, provided users keep the focus on high-level data structures and avoid premature optimization or unnecessary exposure to low-level Dask functionality.
Storage Chunks#
Earth system data is often stored in chunked file formats such as NetCDF4 or Zarr. In this context, storage chunks describe how variables are physically laid out on disk. Each chunk is a contiguous block of data that is read or written in a single I/O operation. Storage chunking is fixed at data creation time and reflects assumptions about how the data will be accessed—for example, reading full time series at individual grid points or loading spatial slices at specific time steps.
For users, storage chunks primarily matter because they define the minimum cost of I/O. Access patterns that align with storage chunks are efficient, while misaligned access can require reading many chunks to assemble a small logical subset of data. Although storage chunking cannot usually be changed without rewriting the dataset, it is essential to be aware of it, as it sets the baseline for performance in any downstream analysis, including those using Dask.
We can inspect the storage chunking of a dataset by providing the chunks={} keyword to xr.open_dataset().
This will create a Dask array that uses the underlying storage chunking directly as Dask chunks:
import intake
import xarray as xr
cat = intake.open_catalog("https://data.nextgems-h2020.eu/online.yaml")
urlpath = cat.ERA5(zoom=6).urlpath
ds = xr.open_dataset(urlpath, chunks={})
ds["2t"].chunks
((24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24,
24),
(16384, 16384, 16384))
In this example, the storage chunking has been configured so that 24 time steps and 16,384 spatial cells must always be loaded at once.
Dask Chunks#
Dask chunks describe how data are partitioned in memory for parallel computation.
When opening a dataset with Dask (for example via xarray’s chunks= argument), users specify how arrays should be split into pieces that can be processed independently.
Each Dask chunk becomes one or more tasks in the Dask task graph, enabling parallel execution across CPU cores or cluster workers.
Unlike storage chunks, Dask chunks are flexible and can be adapted to the analysis at hand.
Ideally, the Dask chunk size is immediately set when creating a Dask array, i.e. when opening a dataset. This avoids an additional rechunking layer in the task graph:
ds_dask = xr.open_dataset(urlpath, chunks={"time": 96})
ds_dask["2t"].chunks
((96, 96, 96, 96, 96, 96, 96, 96, 96, 96, 48), (16384, 16384, 16384))
Warning
The .chunk() method can change the chunking of existing Dask arrays.
Use with care: rechunking is often expensive and can significantly degrade performance.
Prefer choosing suitable chunking when opening the dataset.
Efficient workflows should aim to align Dask chunks with storage chunks, or to combine multiple storage chunks into a single Dask chunk, rather than splitting them further. Good alignment minimizes unnecessary I/O and avoids excessive task overhead. From a practical perspective, users should choose Dask chunks that are large enough to amortize overhead, small enough to fit comfortably in memory, and shaped to match the dominant operations (e.g. chunking along time for temporal reductions). Understanding the distinction—and relationship—between storage chunks and Dask chunks is often the single most important step toward effective and scalable use of Dask with Earth system data.
For this example, we have intentionally chosen a large chunk size for the spatial (cell) dimension.
Additionally, we split the storage chunking in the time dimension.
This is a particularly poor choice, especially when performing temporal reductions, which is reflected in a rather complex task graph:
bad = xr.open_dataset(urlpath, chunks={"cell": -1, "time": 12})
bad["2t"].mean("time").data.visualize()
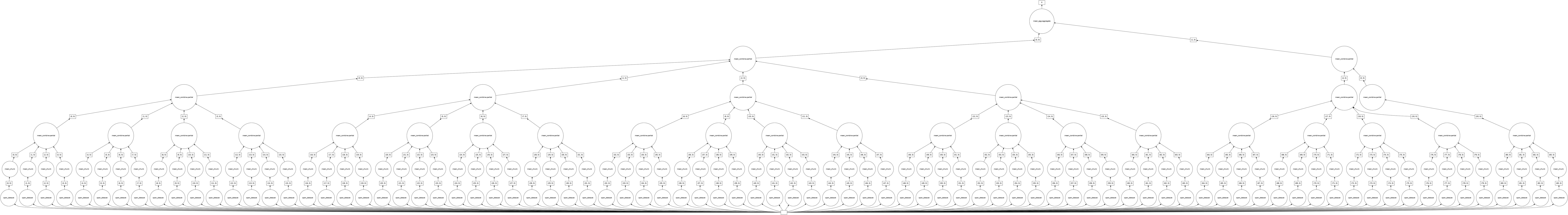
A better pattern would be to define Dask chunks that combine several storage chunks along the time dimension:
good = xr.open_dataset(urlpath, chunks={"time": 256})
good["2t"].mean("time").data.visualize()
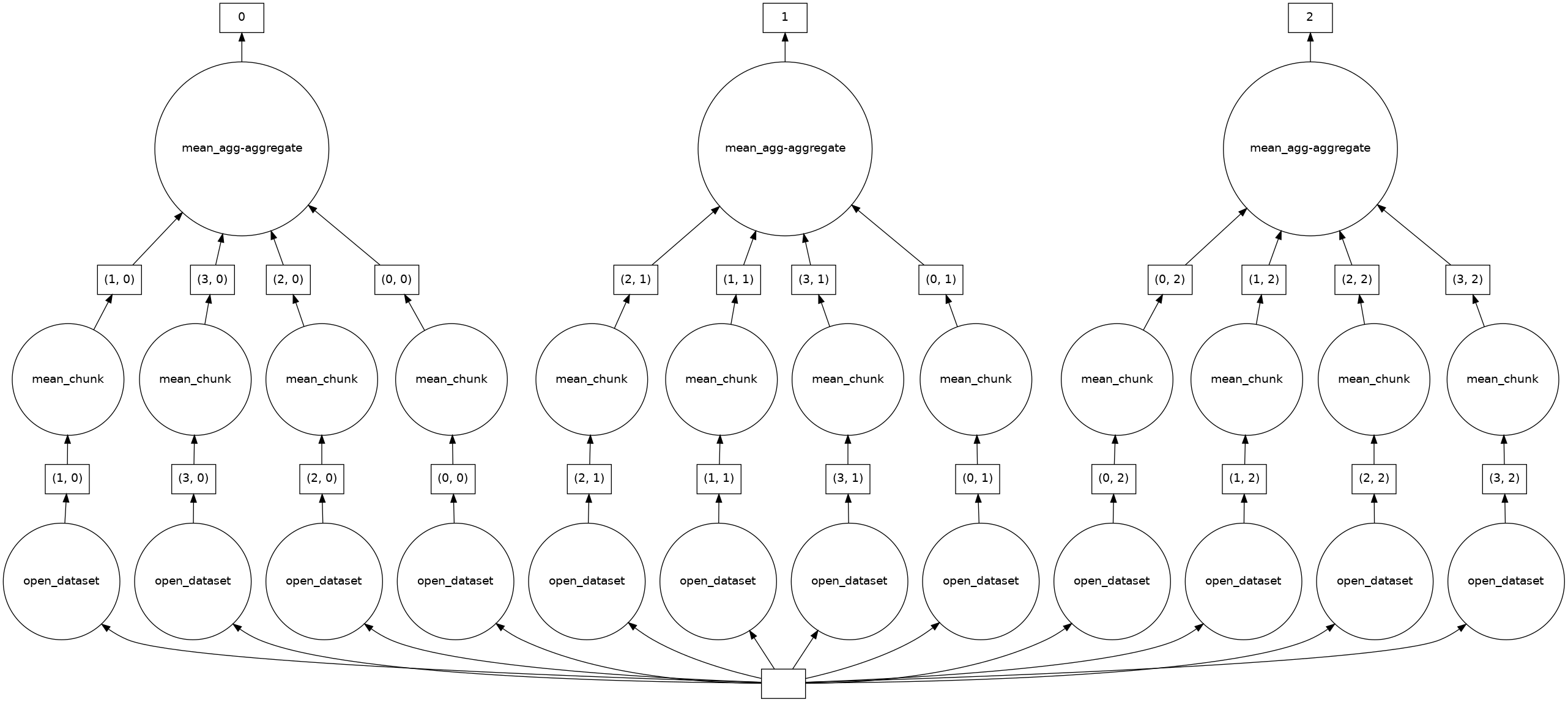
Tip
Technically, the ideal Dask chunk size depends on the amount of memory available to each CPU.
A good rule of thumb is to aim for Dask chunks of around 100 MB.
This size allows for an adequate memory usage buffer, even when many Dask tasks are being executed in parallel.
In recent versions of Dask, you can also set the chunk size to "auto", which allows Dask to determine the size based on storage and analysis.
Neither of the above examples has triggered an execution yet.
This can be achieved by accessing the .values attribute of an Xarray data array, plotting it, or calling the .compute() method manually:
good["2t"].mean("time").compute()
<xarray.DataArray '2t' (cell: 49152)> Size: 197kB
array([299.24762, 299.25766, 299.37292, ..., 299.55402, 299.43484,
299.50705], shape=(49152,), dtype=float32)
Coordinates:
* cell (cell) float32 197kB 0.0 1.0 2.0 ... 4.915e+04 4.915e+04 4.915e+04
crs float32 4B 0.0
lat (cell) float32 197kB 0.5968 1.194 1.194 ... -1.194 -1.194 -0.5968
lon (cell) float32 197kB 45.0 45.7 44.3 45.0 ... 315.7 314.3 315.0
Attributes:
grid_mapping: crs
levtype: surface
long_name: 2 metre temperature
standard_name:
units: KTake-home messages
Dask enables scalable analysis by separating definition from execution.
Storage chunks define how data is laid out on disk and thus set the minimum I/O cost.
Dask chunks define how data is partitioned in memory for parallel computation.
For good performance, Dask chunks should align with (or combine) storage chunks.
Prefer choosing suitable chunking when opening datasets.
